
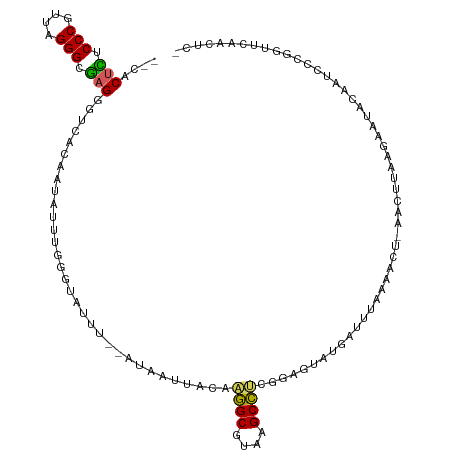
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 704,153 – 704,256 |
| Length | 103 |
| Max. P | 0.988223 |

| Location | 704,153 – 704,256 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.61308 |
| G+C content | 0.40682 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.94 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 704153 103 + 2517507 --CACUCUCCCGUUAGGGAGAGGGACACAAUAUUCAGGUAUUU--AUAAUAACAAGGCGUAAGCCUCGGAGUAUGAUUAAAAACU-AACUUAAGAAUCCGGUCCAAGU-------- --..(((((((....)))))))((((.................--.........((((....))))((((......((((.....-...))))...))))))))....-------- ( -32.40, z-score = -3.96, R) >droSim1.chrU 4000114 110 + 15797150 --CACUCUCCCGUUAGGGCGAGGGUCACAAUAUUUGGGUAUUU--AUAAUUACAAGGCGUAAGCCUCGGAGUAUGAUUUAUAACA-AACUUAAGAAUUCGUUCCCGGUUCAACUC- --..(((.(((....))).)))((..(((((.(((((((..((--((((.(((.((((....))))....)))....))))))..-.))))))).))).))..))..........- ( -27.10, z-score = -1.36, R) >droSec1.super_113 39035 110 + 77858 --CACUCUCCCGUUAGGGCGAGGGUCACAAUAUUUGGGUAUUU--AUAAUUACAAGGCGUAAGCCUCGGAGUAUGAUUUAAAACU-AACUUAAGAAUUCAGUCCCGGUUCAACUC- --...((.(((....))).))(((.(..(((.(((((((..((--(((....(.((((....)))).)...))))).........-.))))))).)))..).)))..........- ( -23.60, z-score = -0.17, R) >droYak2.chr2L_random 3346784 111 - 4064425 --UACUUCCCCGUUAGGGUGAGAGAC-UAAUAUUCGAUUAUUACAAUAAGUACAGAGCGUAAGCUCUGGAGUACAAUUAUUUACUGAACUUAAGUUUACAAUCU-AAUGCAACUC- --..((..(((....)))..))((((-(((((......))))....(((((.((((((....)))))).((((........))))..)))))))))).......-..........- ( -26.20, z-score = -2.18, R) >droAna3.scaffold_13250 1925908 114 + 3535662 CGCUCAAGCCCGUUAGGGUUAGGCUACUGACAAAUAUAAUAUAAAAUAUUACGCGAGCUAAGGCUGCGGAGUUCUGCUUAAGACUAAACU--AGAAUACUACCCGGGCUGUACUUC ......((((((((((..((((((.(((....(((((........))))).(((.(((....)))))).)))...))))))..))))..(--((....)))..))))))....... ( -29.70, z-score = -0.82, R) >consensus __CACUCUCCCGUUAGGGCGAGGGUCACAAUAUUUGGGUAUUU__AUAAUUACAAGGCGUAAGCCUCGGAGUAUGAUUUAAAACU_AACUUAAGAAUACAAUCCCGGUUCAACUC_ ....(((.(((....))).)))................................((((....)))).................................................. (-13.10 = -13.94 + 0.84)

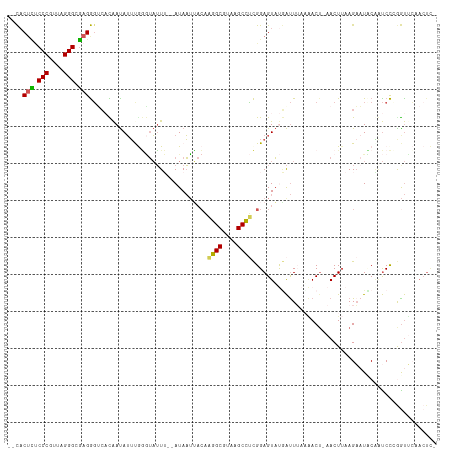
| Location | 704,153 – 704,256 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.61308 |
| G+C content | 0.40682 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -14.84 |
| Energy contribution | -14.76 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 704153 103 - 2517507 --------ACUUGGACCGGAUUCUUAAGUU-AGUUUUUAAUCAUACUCCGAGGCUUACGCCUUGUUAUUAU--AAAUACCUGAAUAUUGUGUCCCUCUCCCUAACGGGAGAGUG-- --------....((((((((((((((((..-....)))))........((((((....)))))).......--........)))).))).))))(((((((....)))))))..-- ( -30.70, z-score = -3.51, R) >droSim1.chrU 4000114 110 - 15797150 -GAGUUGAACCGGGAACGAAUUCUUAAGUU-UGUUAUAAAUCAUACUCCGAGGCUUACGCCUUGUAAUUAU--AAAUACCCAAAUAUUGUGACCCUCGCCCUAACGGGAGAGUG-- -..........((((((((((......)))-)))).............((((((....)))))).......--.....))).............(((.(((....))).)))..-- ( -27.60, z-score = -2.06, R) >droSec1.super_113 39035 110 - 77858 -GAGUUGAACCGGGACUGAAUUCUUAAGUU-AGUUUUAAAUCAUACUCCGAGGCUUACGCCUUGUAAUUAU--AAAUACCCAAAUAUUGUGACCCUCGCCCUAACGGGAGAGUG-- -(((((((...((((((((.........))-))))))...))).))))((((((....)))))).......--.....................(((.(((....))).)))..-- ( -26.60, z-score = -1.63, R) >droYak2.chr2L_random 3346784 111 + 4064425 -GAGUUGCAUU-AGAUUGUAAACUUAAGUUCAGUAAAUAAUUGUACUCCAGAGCUUACGCUCUGUACUUAUUGUAAUAAUCGAAUAUUA-GUCUCUCACCCUAACGGGGAAGUA-- -(((..(((((-.((((((.....(((((..((((........)))).((((((....)))))).))))).....)))))).)))....-))..))).(((....)))......-- ( -25.20, z-score = -1.60, R) >droAna3.scaffold_13250 1925908 114 - 3535662 GAAGUACAGCCCGGGUAGUAUUCU--AGUUUAGUCUUAAGCAGAACUCCGCAGCCUUAGCUCGCGUAAUAUUUUAUAUUAUAUUUGUCAGUAGCCUAACCCUAACGGGCUUGAGCG ...((.(((((((((.(((..(((--.(((((....)))))))))))))(((((....))).))(((((((...))))))).......................))))).)).)). ( -25.30, z-score = 0.36, R) >consensus _GAGUUGAACCGGGACCGAAUUCUUAAGUU_AGUUUUAAAUCAUACUCCGAGGCUUACGCCUUGUAAUUAU__AAAUACCCAAAUAUUGUGACCCUCACCCUAACGGGAGAGUG__ ................................................((((((....))))))..............................(((.(((....))).))).... (-14.84 = -14.76 + -0.08)


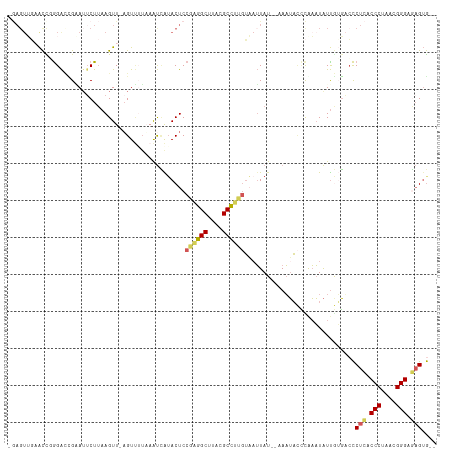
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:56 2011