| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 687,788 – 687,870 |
| Length | 82 |
| Max. P | 0.637714 |

| Location | 687,788 – 687,870 |
|---|---|
| Length | 82 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 63.97 |
| Shannon entropy | 0.68546 |
| G+C content | 0.51634 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -10.82 |
| Energy contribution | -9.89 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.22 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
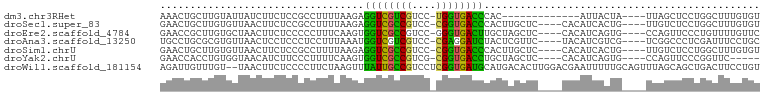
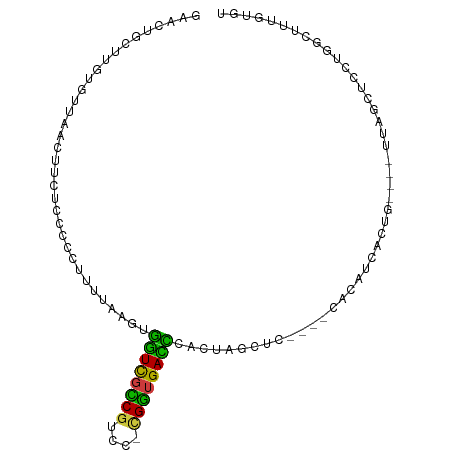
>dm3.chr3RHet 687788 82 + 2517507 AAACUGCUUGUAUUAUCUUCUCCGCCUUUUAAGAGGUCGUCGUCC-UGGUGACCCAC-------------AUUACUA----UUAGCUCCUGGCUUUGUGU ........((..(((((..(..((((((....)))).))..)...-.)))))..)).-------------..(((..----..(((.....)))..))). ( -11.70, z-score = 1.04, R) >droSec1.super_83 11412 91 - 109685 GAACUGCUUGUGUUAACUUCUCCGCCUUUUAAGAGGUCGCCGUCC-CGGUGACCCACUUGCUC----CACAUCACUG----UUGUCUCCUGGCUUUGUGU .....((..(((......(((..........)))((((((((...-)))))))))))..))..----.....(((..----..(((....)))...))). ( -21.20, z-score = -0.92, R) >droEre2.scaffold_4784 1233528 91 - 25762168 GAACCGCUUGUGCUAACUUCUCCCCCUUUCAAGUGGUCGCCGUCC-GGGUGACUUGCUAGCUC----CACAUCAGUG----CCAGUUCCCUGUUUUGUUC ((.(((((((...................)))))))))((.(..(-(((..(((.((..(((.----......))))----).)))..))))..).)).. ( -20.41, z-score = -0.58, R) >droAna3.scaffold_13250 1919502 91 + 3535662 UGCCUGCGCGUGUUAACUCCUCCCUCCUUUAAAUGGUCGUCGUCC-CGAGGAUCUACUCGUUC----UACAUCGUCG----UCGGCCCUCGAUUUCCUGC .(((.((((((((.(((.....(..((.......))..)......-.(((......)))))).----.))).)).))----).))).............. ( -17.10, z-score = 0.19, R) >droSim1.chrU 8715982 91 - 15797150 GAACUGCUUGUGUUAACUUCUCCGCCUUUUAAGAGGUCGCCGUCC-CGGUGACCCACUUGCUC----CACAUCACUG----UUGUCUCCUGGCUUUGUGU .....((..(((......(((..........)))((((((((...-)))))))))))..))..----.....(((..----..(((....)))...))). ( -21.20, z-score = -0.92, R) >droYak2.chrU 1739077 86 + 28119190 GAACCACCUGUGGUAACAUCUUCCCUUUUCAAGUGGUCGCCGUCG-CGGUGACCUGCUAGCUC----CACAUCAGUG----CCAGUUCCCGGUUC----- (((((..........................(((((((((((...-)))))))).)))((((.----(((....)))----..))))...)))))----- ( -22.60, z-score = -0.84, R) >droWil1.scaffold_181154 1014067 98 + 4610121 AGAUUGUUUGU--UAACUUCUCCCCUUCUAAGUUUAUUGCCGUCCUCGGUGAUGCAUGACACUUGGACGAAUUUUUGCAGUUUAGCAGCUGACUUCCUGU (((((((....--.((.((((((........((.(((..(((....)))..))).)).......))).))).))..))))))).((((........)))) ( -17.36, z-score = 0.50, R) >consensus GAACUGCUUGUGUUAACUUCUCCCCCUUUUAAGUGGUCGCCGUCC_CGGUGACCCACUAGCUC____CACAUCACUG____UUAGCUCCUGGCUUUGUGU ..................................((((((((....)))))))).............................................. (-10.82 = -9.89 + -0.93)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:54 2011