| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 683,752 – 683,814 |
| Length | 62 |
| Max. P | 0.989981 |

| Location | 683,752 – 683,814 |
|---|---|
| Length | 62 |
| Sequences | 9 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 65.40 |
| Shannon entropy | 0.68657 |
| G+C content | 0.43156 |
| Mean single sequence MFE | -12.13 |
| Consensus MFE | -6.87 |
| Energy contribution | -6.34 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.989981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
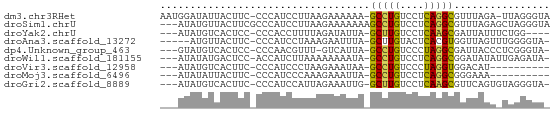
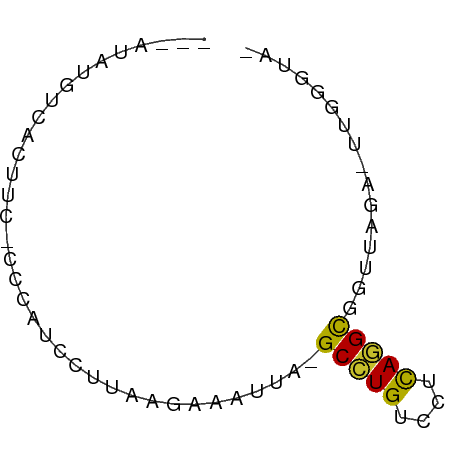
>dm3.chr3RHet 683752 62 + 2517507 AAUGGAUAUUACUUC-CCCAUCCUUAAGAAAAAA-GCCUGUCCUCAGGCGUUUAGA-UUAGGGUA ...(((.......))-)....((((((.(((...-(((((....))))).)))...-)))))).. ( -16.10, z-score = -1.68, R) >droSim1.chrU 2589943 62 + 15797150 ---AUAUGUUACUUCGCCCAUCCUUAAGAAAAAAAGCCUGUCCUCAGGCGUUUAGAGCUAGGGUA ---............((((...(((.(((......(((((....))))).))).)))...)))). ( -14.80, z-score = -1.19, R) >droYak2.chrU 22661494 56 - 28119190 ---AUAUGUCACUCC-CCCACCUUUUAGAUAUUA-GCUUGUCCUCAAGCGAUUAUUUCUGG---- ---............-....((....(((((...-(((((....)))))...)))))..))---- ( -8.60, z-score = -1.87, R) >droAna3.scaffold_13272 57971 57 - 529493 -----AUGUUACUUC-CCCAUCCUAAAGAAUUUA-GCUUGUACUCACGUGGUUAGUUUGGGGUA- -----.........(-((((..((((........-((.((....)).))..))))..)))))..- ( -11.90, z-score = -0.52, R) >dp4.Unknown_group_463 4742 58 - 6425 ---GUAUGUCACUCC-CCCAACGUUU-GUCAUUA-GCCUGUCCCUAGGCGAUUACCCUCGGGUA- ---............-(((.......-.......-(((((....)))))((......)))))..- ( -10.60, z-score = -0.07, R) >droWil1.scaffold_181155 1006375 59 - 3442787 ---AUAUAUGACUCC-ACCAUCUUAAAAAAAAUA-GCCUGUCCUCAGGCGGAUAUAUUGAGAUA- ---............-...(((((((........-(((((....))))).......))))))).- ( -13.06, z-score = -2.45, R) >droVir3.scaffold_12958 3344168 50 + 3547706 ---AUAUGUCACUUC-CCCAUCCCUAAGAAAUAA-GCCUGUCCCUAGGUGGACAU---------- ---..(((((.....-....((.....)).....-(((((....))))).)))))---------- ( -7.10, z-score = -0.03, R) >droMoj3.scaffold_6496 2333102 50 - 26866924 ---AUAUAUUACUUC-CCCAUCCCAAAGAAAUUA-GCCUGUCCUCAGGCGGGAAA---------- ---............-....((((.((....)).-(((((....)))))))))..---------- ( -14.60, z-score = -3.10, R) >droGri2.scaffold_8889 8465 59 + 17201 ---AUAUGUCACUUC-CCCAUCCAUUAGAAAUUG-GCUUGUCCUCAAGCGUUCAGUGUAGGGUA- ---............-(((...((((.(((....-(((((....))))).)))))))..)))..- ( -12.40, z-score = -1.18, R) >consensus ___AUAUGUCACUUC_CCCAUCCUUAAGAAAUUA_GCCUGUCCUCAGGCGGUUAGA_UUGGGUA_ ...................................(((((....)))))................ ( -6.87 = -6.34 + -0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:53 2011