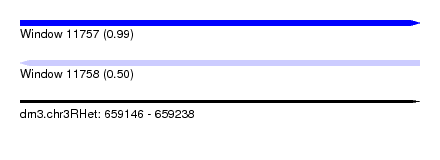
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 659,146 – 659,238 |
| Length | 92 |
| Max. P | 0.993553 |

| Location | 659,146 – 659,238 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.98 |
| Shannon entropy | 0.39766 |
| G+C content | 0.50191 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -20.51 |
| Energy contribution | -21.58 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
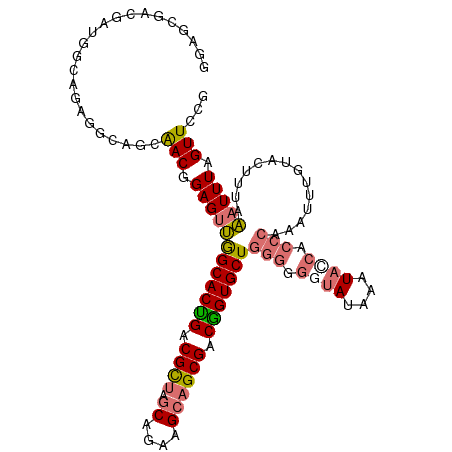
>dm3.chr3RHet 659146 92 + 2517507 ------------------GGAGCGAC-GAGUUGGCACUGACGUUAGCAGAAGCAGCGACAGUGCUGGGGGGUAUAAAUAUCACCCAAUUUUGUACUUUAAAUUUAGUUCCG ------------------(((((...-...(..((((((.((((.((....)))))).))))))..)((((((((((...........)))))))))).......))))). ( -31.90, z-score = -3.62, R) >droSim1.chrU 2292476 104 - 15797150 GGAGCGACGAUGGCAGAGUCAGCAACGGAGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCU-------AUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCG ((((((((.........)))....(((((((((((((((.((((.((....)))))).)))))).-------............)))))))))............))))). ( -31.81, z-score = -2.22, R) >droSec1.super_42 220670 111 - 298440 GGAGCGACGAUGGCAGAGCCAGCAACUGAGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGGGGUAUAAAUACCACCCAAAUAUGUACUUUAAAUUUAGUUCCG (((((..((....(((.((((((......)))))).))).((((.((....)))))).))((((((((.((((....)))).)))).....))))..........))))). ( -41.40, z-score = -3.64, R) >droYak2.chrU 17377185 110 + 28119190 GGAGCAACGAGGGCAGAGGCAGCGACGGAGCCAGCACCGACGCCAACAGAGGCAGCGACGGUGCUGGGAGGUAUAAAUAACACCCAAAU-UGUAAAAUAGCUUUAGUUCAA .((((..((...((....))..))..(((((((((((((.(((...........))).))))))))((..((.......))..))....-.........))))).)))).. ( -33.20, z-score = -1.96, R) >consensus GGAGCGACGAUGGCAGAGGCAGCAACGGAGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGGGGUAUAAAUACCACCCAAAUUUGUACUUUAAAUUUAGUUCCG .......................(((.((((((((((((.((((.((....)))))).)))))))(((.((((....)))).)))..............))))).)))... (-20.51 = -21.58 + 1.06)

| Location | 659,146 – 659,238 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.98 |
| Shannon entropy | 0.39766 |
| G+C content | 0.50191 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
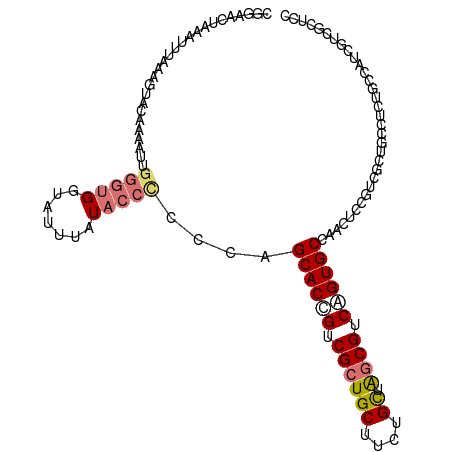
>dm3.chr3RHet 659146 92 - 2517507 CGGAACUAAAUUUAAAGUACAAAAUUGGGUGAUAUUUAUACCCCCCAGCACUGUCGCUGCUUCUGCUAACGUCAGUGCCAACUC-GUCGCUCC------------------ .((.(((........)))........(((((.......))))).)).((((((.((..((....))...)).))))))......-........------------------ ( -20.50, z-score = -1.42, R) >droSim1.chrU 2292476 104 + 15797150 CGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAU-------AGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACUCCGUUGCUGACUCUGCCAUCGUCGCUCC .((((((........))).........(((((((.....-------.((((.(.((((((....)).)))).).))))((((...)))).......))))))).....))) ( -25.80, z-score = -0.56, R) >droSec1.super_42 220670 111 + 298440 CGGAACUAAAUUUAAAGUACAUAUUUGGGUGGUAUUUAUACCCCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACUCAGUUGCUGGCUCUGCCAUCGUCGCUCC .(.((((..................((((.((((....)))).))))((((.(.((((((....)).)))).).))))......)))).)((((...)))).......... ( -32.10, z-score = -1.67, R) >droYak2.chrU 17377185 110 - 28119190 UUGAACUAAAGCUAUUUUACA-AUUUGGGUGUUAUUUAUACCUCCCAGCACCGUCGCUGCCUCUGUUGGCGUCGGUGCUGGCUCCGUCGCUGCCUCUGCCCUCGUUGCUCC .........(((.........-....((((((.....)))))).(((((((((.((((.........)))).))))))))).......))).................... ( -32.40, z-score = -2.23, R) >consensus CGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUACCCCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACUCCGUCGCUGCCUCUGCCAUCGUCGCUCC ..........................(((((.......)))))....((((((.((((((....)).)))).))))))................................. (-16.20 = -17.32 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:52 2011