
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 612,469 – 612,524 |
| Length | 55 |
| Max. P | 0.941158 |

| Location | 612,469 – 612,524 |
|---|---|
| Length | 55 |
| Sequences | 4 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 73.05 |
| Shannon entropy | 0.44608 |
| G+C content | 0.44310 |
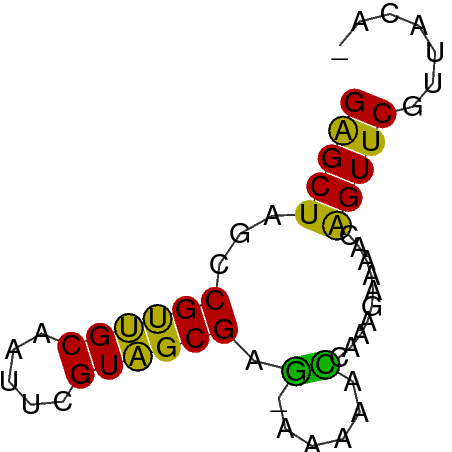
| Mean single sequence MFE | -13.12 |
| Consensus MFE | -8.14 |
| Energy contribution | -7.33 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941158 |
| Prediction | RNA |
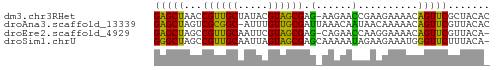
Download alignment: ClustalW | MAF
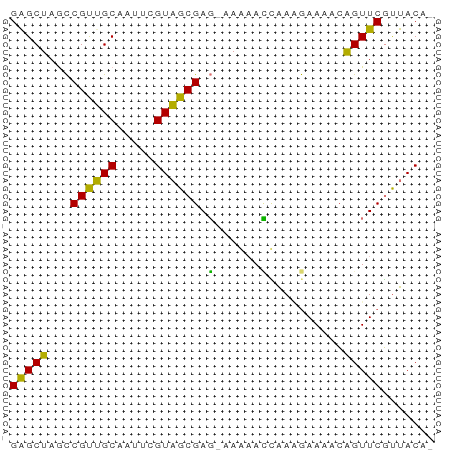
>dm3.chr3RHet 612469 55 - 2517507 GAGCUAACCGUUGCUAUACGUAGCGAG-AAGAACCGAAGAAAACAGUUCGCUACAC .(((........)))....((((((((-..(............)..)))))))).. ( -12.50, z-score = -1.94, R) >droAna3.scaffold_13339 1531881 55 - 4599533 GAGCUAGUCGCGGC-AUUUGUUGCGAUUAAACAAUAACAAAAACAGUUCGUUACAC ((((((((((((((-....)))))))))................)))))....... ( -13.69, z-score = -2.39, R) >droEre2.scaffold_4929 1255907 54 + 26641161 GAGCUAGCCGUUGCAAUUCGUAGCGAG-CAGAACCAAGGAAAACAGUUCGUUACA- (((((.((((((((.....)))))).)-)....(....).....)))))......- ( -13.80, z-score = -1.87, R) >droSim1.chrU 4806313 55 + 15797150 GGGCUAGCCGUUGCAAUUAGUAGCGAGCAAAAAUAGAAGAAAUGGGUUCUUUACA- ......((((((((.....)))))).)).......((((((.....))))))...- ( -12.50, z-score = -1.09, R) >consensus GAGCUAGCCGUUGCAAUUCGUAGCGAG_AAAAACCAAAGAAAACAGUUCGUUACA_ (((((...((((((.....))))))........(....).....)))))....... ( -8.14 = -7.33 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:50 2011