| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 610,049 – 610,143 |
| Length | 94 |
| Max. P | 0.952691 |

| Location | 610,049 – 610,143 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 60.96 |
| Shannon entropy | 0.53291 |
| G+C content | 0.39168 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -11.84 |
| Energy contribution | -13.19 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
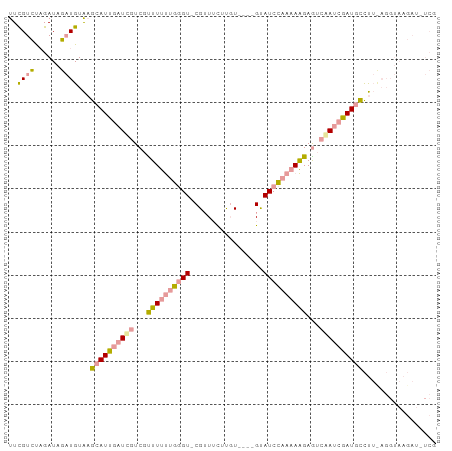
>dm3.chr3RHet 610049 94 + 2517507 UUCGUCCAGAUAGAUGUAAGCAUUGAUCGUCGUUUUUUGGGU-CGAUUCUUGU----GUAUCCAAAAAGAGUCAAUCGAUGCCUUUAGGUAAGAUUUCG ..((((......))))...((((((((.(.(.((((((((((-((.......)----).)))))))))).).).))))))))................. ( -20.70, z-score = -1.00, R) >droSec1.super_46 224355 94 + 322281 UUUGUCUAGAUAGAUGUAAGCAUCGAUCGUCGCUUUUCGGGU-CGUUACUUGU----GCAUCCAAAAAGAGUCGAUCGAUGCCUUCAGGUAAGAUAUCG ..(((((..((...((...((((((((((.(.(((((.((((-.((.......----)))))).))))).).))))))))))...)).)).)))))... ( -30.50, z-score = -3.21, R) >droYak2.chrX 21227154 84 + 21770863 UUCGUGUUGAUCCUUGUGUGUAUUGAUUGUCGUUUCCUGUGUGCGUUUUUCUUAGUUGUAUCUAAUAGGAAGAAAAAGAUUUUC--------------- ........................((..(((.(((((((((((((...........)))))...)))))))).....)))..))--------------- ( -11.20, z-score = -0.62, R) >consensus UUCGUCUAGAUAGAUGUAAGCAUUGAUCGUCGUUUUUUGGGU_CGUUUCUUGU____GUAUCCAAAAAGAGUCAAUCGAUGCCUU_AGGUAAGAU_UCG ..((((......))))...((((((((((...((((((((((.(.............).))))))))))...))))))))))................. (-11.84 = -13.19 + 1.35)


| Location | 610,049 – 610,143 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 60.96 |
| Shannon entropy | 0.53291 |
| G+C content | 0.39168 |
| Mean single sequence MFE | -17.13 |
| Consensus MFE | -5.78 |
| Energy contribution | -7.57 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
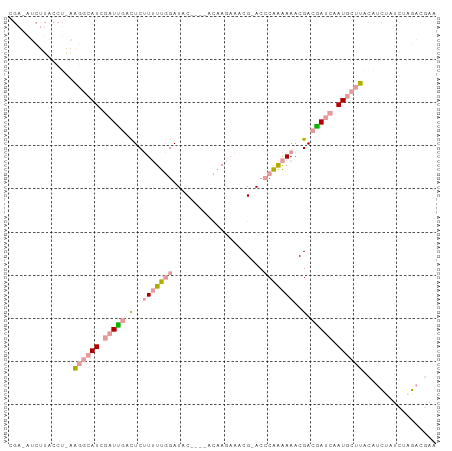
>dm3.chr3RHet 610049 94 - 2517507 CGAAAUCUUACCUAAAGGCAUCGAUUGACUCUUUUUGGAUAC----ACAAGAAUCG-ACCCAAAAAACGACGAUCAAUGCUUACAUCUAUCUGGACGAA ((.............((((((.(((((.(..(((((((....----..........-..)))))))..).))))).))))))...(((....))))).. ( -15.89, z-score = -1.33, R) >droSec1.super_46 224355 94 - 322281 CGAUAUCUUACCUGAAGGCAUCGAUCGACUCUUUUUGGAUGC----ACAAGUAACG-ACCCGAAAAGCGACGAUCGAUGCUUACAUCUAUCUAGACAAA .((((((......))((((((((((((.(.((((((((.(((----....)))...-..)))))))).).)))))))))))).....))))........ ( -29.10, z-score = -4.47, R) >droYak2.chrX 21227154 84 - 21770863 ---------------GAAAAUCUUUUUCUUCCUAUUAGAUACAACUAAGAAAAACGCACACAGGAAACGACAAUCAAUACACACAAGGAUCAACACGAA ---------------((((.....)))).((((.((((......))))..............(....).................)))).......... ( -6.40, z-score = -1.20, R) >consensus CGA_AUCUUACCU_AAGGCAUCGAUUGACUCUUUUUGGAUAC____ACAAGAAACG_ACCCAAAAAACGACGAUCAAUGCUUACAUCUAUCUAGACGAA ...............((((((.(((((.(..(((((((.....................)))))))..).))))).))))))................. ( -5.78 = -7.57 + 1.79)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:48 2011