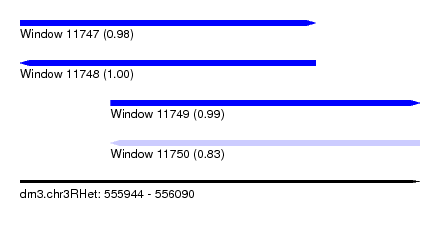
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 555,944 – 556,090 |
| Length | 146 |
| Max. P | 0.999227 |

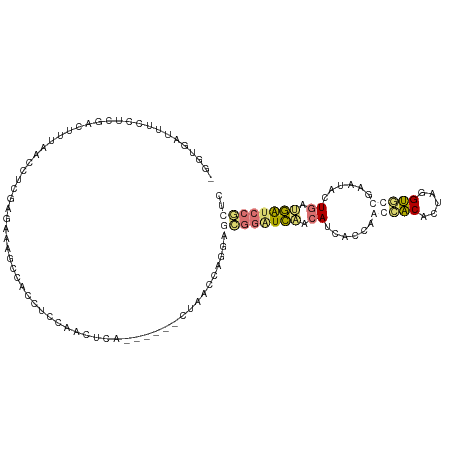
| Location | 555,944 – 556,052 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 54.03 |
| Shannon entropy | 0.74994 |
| G+C content | 0.53066 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -9.31 |
| Energy contribution | -10.75 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 555944 108 + 2517507 GGGAGAGCCUUUGCUCUUCAACCUUGCAACAAAAGCAUCUAUCUCAGAGGAUUCAACCGGUGGUGGAUUCUCAUCAGGAAUGACAGUGGGUGCCGAAAACUCCGAGAG----- .(((((((....)))))))...((((........((((((((....(((((((((.((...))))))))))).(((....)))..)))))))).........))))..----- ( -32.03, z-score = -0.43, R) >droAna3.scaffold_8169 520 106 - 1573 -GGCGCUUUCCCAGAGCGAAGCUUAGUGAAAGCCCACUGAAGCUCG------CUCAUCAGGAGGGGAUCAGCAUCACCCACUGCAUCAGGCACUGAAUGCUGCCGAUCCUCCC -............((((((.((((((((......))))).))))))------)))....((.(((((((.(((.((..((.(((.....))).))..)).))).))))))))) ( -41.40, z-score = -2.37, R) >droVir3.scaffold_12943 155303 106 - 194972 -GGUGACUUCCUCGACUUUAACCUCGAGAAAGCCACCUCCAACUCA------CUAUCCAGAAGCGGAUCAACAGGCCUAACCACACUAGGUGGUGAGUCCUGAUGAUCCGCUC -((((.(((.(((((........))))).))).)))).........------.........(((((((((.((((.((.(((((.....))))).)).)))).))))))))). ( -45.30, z-score = -6.28, R) >droGri2.scaffold_15056 5481 106 + 38174 -GGUGAUUUCCUCGACUUUAACAUCGAGAAAGCCACCUCCAAUUCA------CUAUCCAGGAGCGGGUCAACAUCACCAACCACACUCGGUGGCAAGUUCUGAUGAUCCGGUC -((((..((.(((((........))))).))..)))).........------........((.(((((((.((..((...((((.....))))...))..)).))))))).)) ( -30.60, z-score = -1.70, R) >consensus _GGUGAUUUCCUCGACUUUAACCUCGAGAAAGCCACCUCCAACUCA______CUAACCAGGAGCGGAUCAACAUCACCAACCACACUAGGUGCCGAAUACUGAUGAUCCGCUC ..............................................................((((((((.((......(((((.....)))))......)).)))))))).. ( -9.31 = -10.75 + 1.44)

| Location | 555,944 – 556,052 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 54.03 |
| Shannon entropy | 0.74994 |
| G+C content | 0.53066 |
| Mean single sequence MFE | -43.73 |
| Consensus MFE | -14.54 |
| Energy contribution | -19.98 |
| Covariance contribution | 5.44 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 555944 108 - 2517507 -----CUCUCGGAGUUUUCGGCACCCACUGUCAUUCCUGAUGAGAAUCCACCACCGGUUGAAUCCUCUGAGAUAGAUGCUUUUGUUGCAAGGUUGAAGAGCAAAGGCUCUCCC -----.((((((((..((((((......((..(((((....).)))).))......))))))..)))))))).(((.(((((((((.((....))...))))))))))))... ( -30.60, z-score = -0.49, R) >droAna3.scaffold_8169 520 106 + 1573 GGGAGGAUCGGCAGCAUUCAGUGCCUGAUGCAGUGGGUGAUGCUGAUCCCCUCCUGAUGAG------CGAGCUUCAGUGGGCUUUCACUAAGCUUCGCUCUGGGAAAGCGCC- .((.(((((((((.((((((.(((.....))).)))))).)))))))))))((((...(((------(((((((.(((((....))))))))).)))))).)))).......- ( -52.50, z-score = -3.57, R) >droVir3.scaffold_12943 155303 106 + 194972 GAGCGGAUCAUCAGGACUCACCACCUAGUGUGGUUAGGCCUGUUGAUCCGCUUCUGGAUAG------UGAGUUGGAGGUGGCUUUCUCGAGGUUAAAGUCGAGGAAGUCACC- .(((((((((.((((.((.(((((.....))))).)).)))).)))))))))((..(....------....)..))((((((((((((((........))))))))))))))- ( -53.30, z-score = -5.85, R) >droGri2.scaffold_15056 5481 106 - 38174 GACCGGAUCAUCAGAACUUGCCACCGAGUGUGGUUGGUGAUGUUGACCCGCUCCUGGAUAG------UGAAUUGGAGGUGGCUUUCUCGAUGUUAAAGUCGAGGAAAUCACC- ...(((.(((.((..(((.(((((.....))))).)))..)).))).)))((((.......------......))))(((..(((((((((......)))))))))..))).- ( -38.52, z-score = -2.49, R) >consensus GAGCGGAUCAGCAGAACUCACCACCUAGUGUGGUUGGUGAUGUUGAUCCGCUCCUGGAUAG______UGAGUUGGAGGUGGCUUUCUCGAGGUUAAAGUCGAGGAAAUCACC_ ((((((((((.((.((((.(((((.....))))).)))).)).)))))))))).......................(((((.((((((((........)))))))).))))). (-14.54 = -19.98 + 5.44)


| Location | 555,977 – 556,090 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.14 |
| Shannon entropy | 0.70368 |
| G+C content | 0.49994 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -12.32 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 555977 113 + 2517507 AGCAUCUAUCUCAGAGGAUUCAACCGGUGGUGGAUUCUCAUCAGGAAUGACAGUGGGUGCCGAAAACU-----CCGAGAGUGGAAAAAAGCUCCGGAGA-AAAACACCC-GCGCAGUUGU .((..........(((((((((.((...))))))))))).............(((((((.......((-----(((.((((........))))))))).-....)))))-))))...... ( -38.80, z-score = -2.19, R) >droAna3.scaffold_8169 549 116 - 1573 ---AGCCCACUGAAGCUCGCUCAUCAGGAGGGGAUCAGCAUCACCCACUGCAUCAGGCACUGAAUGCUGCCGAUCCUCCCUAGGAAAGAAUUCGGACAA-AAGACAUUCCACACAAUCCU ---......(((((..((..((...(((.(((((((.(((.((..((.(((.....))).))..)).))).))))))))))..))..)).)))))....-.................... ( -32.20, z-score = -1.05, R) >droVir3.scaffold_12943 155332 113 - 194972 ---AGCCACCUCCAACUCACUAUCCAGAAGCGGAUCAACAGGCCUAACCACACUAGGUGGUGAGUCCUGAUGAUCCGCUCUGGGGAAGAAUUCAUUCAACAAGACAUUC-AUGCAAU--- ---.((................((((((.((((((((.((((.((.(((((.....))))).)).)))).))))))))))))))...((((((.........)).))))-..))...--- ( -41.80, z-score = -4.55, R) >droGri2.scaffold_15056 5510 116 + 38174 ---AGCCACCUCCAAUUCACUAUCCAGGAGCGGGUCAACAUCACCAACCACACUCGGUGGCAAGUUCUGAUGAUCCGGUCUAGGGAAGAACUCAUUCAAUAAGACAUUC-AUACAGUCAC ---................((.(((.(((.(((((((.((..((...((((.....))))...))..)).))))))).))).))).))..............(((....-.....))).. ( -28.00, z-score = -1.28, R) >consensus ___AGCCACCUCAAACUCACUAACCAGGAGCGGAUCAACAUCACCAACCACACUAGGUGCCGAAUACUGAUGAUCCGCUCUAGGAAAGAACUCAGUCAA_AAGACAUUC_ACACAAUC_U .......................(((((.((((((((.((......(((((.....)))))......)).)))))))))))))..................................... (-12.32 = -14.20 + 1.88)


| Location | 555,977 – 556,090 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.14 |
| Shannon entropy | 0.70368 |
| G+C content | 0.49994 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 555977 113 - 2517507 ACAACUGCGC-GGGUGUUUU-UCUCCGGAGCUUUUUUCCACUCUCGG-----AGUUUUCGGCACCCACUGUCAUUCCUGAUGAGAAUCCACCACCGGUUGAAUCCUCUGAGAUAGAUGCU ........((-(((((((..-.((((((((..........))).)))-----)).....))))))).(((((..((..((.(.((.((.(((...))).)).))))).)))))))..)). ( -31.70, z-score = -0.79, R) >droAna3.scaffold_8169 549 116 + 1573 AGGAUUGUGUGGAAUGUCUU-UUGUCCGAAUUCUUUCCUAGGGAGGAUCGGCAGCAUUCAGUGCCUGAUGCAGUGGGUGAUGCUGAUCCCCUCCUGAUGAGCGAGCUUCAGUGGGCU--- ((((..(..((((.......-...))))....)..)))).(((.(((((((((.((((((.(((.....))).)))))).))))))))))))(((..((((.....))))..)))..--- ( -41.30, z-score = -0.75, R) >droVir3.scaffold_12943 155332 113 + 194972 ---AUUGCAU-GAAUGUCUUGUUGAAUGAAUUCUUCCCCAGAGCGGAUCAUCAGGACUCACCACCUAGUGUGGUUAGGCCUGUUGAUCCGCUUCUGGAUAGUGAGUUGGAGGUGGCU--- ---...((..-..((.(((.........(((((....(((((((((((((.((((.((.(((((.....))))).)).)))).)))))))).))))).....)))))))).)).)).--- ( -43.20, z-score = -2.91, R) >droGri2.scaffold_15056 5510 116 - 38174 GUGACUGUAU-GAAUGUCUUAUUGAAUGAGUUCUUCCCUAGACCGGAUCAUCAGAACUUGCCACCGAGUGUGGUUGGUGAUGUUGACCCGCUCCUGGAUAGUGAAUUGGAGGUGGCU--- ....(((.((-((..((((....(((....)))......))))....))))))).....(((((((((((.(((..(.....)..))))))))(..(........)..).)))))).--- ( -32.60, z-score = -0.78, R) >consensus A_GACUGCAU_GAAUGUCUU_UUGAAUGAAUUCUUCCCCAGACCGGAUCAUCAGAACUCACCACCUAGUGUGGUUGGUGAUGUUGAUCCGCUCCUGGAUAGUGAGUUGGAGGUGGCU___ ...........((((.((.........))))))....(((((((((((((.((.((((.(((((.....))))).)))).)).)))))))).)))))....................... (-14.45 = -15.95 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:46 2011