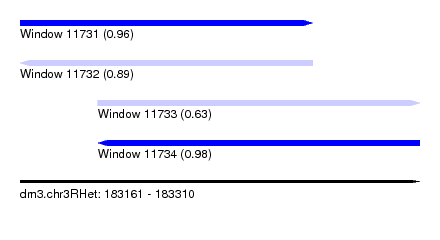
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 183,161 – 183,310 |
| Length | 149 |
| Max. P | 0.983240 |

| Location | 183,161 – 183,270 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 55.08 |
| Shannon entropy | 0.65210 |
| G+C content | 0.57247 |
| Mean single sequence MFE | -47.60 |
| Consensus MFE | -11.96 |
| Energy contribution | -12.31 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 183161 109 + 2517507 ---------ACGGGCUAAAAACCCAUUACCGCAACUUGGUCAUCCUGGGGCCCGGGAUGACCCUAACCGUAAGGGGCUGAGCGGGGCCAUAACGAAGUGGUAUCUUCGUUACCUUCAG ---------..(((.......)))....((((.....(((((((((((...)))))))))))....((....))......))))((...((((((((......))))))))))..... ( -43.50, z-score = -1.99, R) >droGri2.scaffold_14822 128551 118 - 1097346 GCGCCCAAAGAGGAAGGGAGAGCUCCUGCCACCAAGGGGUCAACCCGGUGGGCUGGAUGACCCGAACCGGAGAAAUCUCUGCGGUUCAAGGGUCAAGUGGUACCUCCGGUACCUCCAG ..((((.....((..((((....)))).))(((..(((.....))))))))))((((((((((((((((.(((....))).))))))..))))))...((((((...)))))))))). ( -55.60, z-score = -2.65, R) >droVir3.scaffold_12823 181967 115 - 2474545 GCGUCAGAAACGCGAACCGAAUAUAACGUCACC---GGGACAACCCGGUGGCAAGGGUGAUCCAAACAGAAGAAACCUGAGCGGAUCGCGCACCAAAUGGUAUGUGCGCUACCUCAAA ((((.....))))..............((((((---(((....)))))))))..(((((((((...(((.......)))...)))))((((((..........)))))).)))).... ( -43.70, z-score = -3.76, R) >consensus GCG_C__AAACGGGAAAAAAACCUAAUGCCACCA__GGGUCAACCCGGUGGCCAGGAUGACCCAAACCGAAGAAACCUGAGCGGAUCAAGAACCAAGUGGUAUCUCCGCUACCUCCAG ...........(((............((((((....((((((.((.(.....).)).))))))...(((............)))............))))))..)))........... (-11.96 = -12.31 + 0.35)

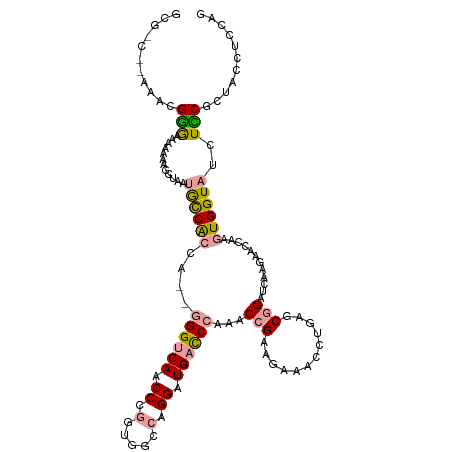
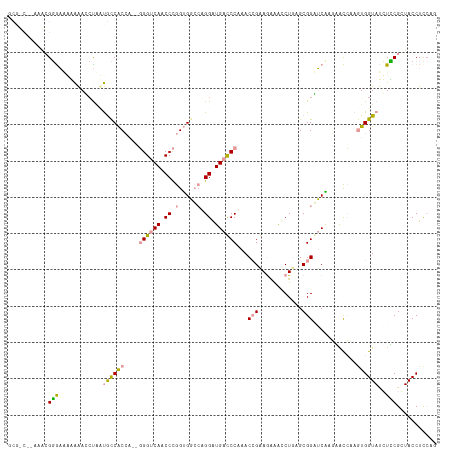
| Location | 183,161 – 183,270 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 55.08 |
| Shannon entropy | 0.65210 |
| G+C content | 0.57247 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -12.99 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 183161 109 - 2517507 CUGAAGGUAACGAAGAUACCACUUCGUUAUGGCCCCGCUCAGCCCCUUACGGUUAGGGUCAUCCCGGGCCCCAGGAUGACCAAGUUGCGGUAAUGGGUUUUUAGCCCGU--------- ......(((((((((......)))))))))((....(((.(((((.((((.((...((((((((.((...)).)))))))).....)).)))).)))))...)))))..--------- ( -41.40, z-score = -1.78, R) >droGri2.scaffold_14822 128551 118 + 1097346 CUGGAGGUACCGGAGGUACCACUUGACCCUUGAACCGCAGAGAUUUCUCCGGUUCGGGUCAUCCAGCCCACCGGGUUGACCCCUUGGUGGCAGGAGCUCUCCCUUCCUCUUUGGGCGC .((((((((((...))))))...((((((..((((((..(((....)))))))))))))))))))((((((((((....)))...)))((.(((.......))).)).....)))).. ( -52.00, z-score = -1.87, R) >droVir3.scaffold_12823 181967 115 + 2474545 UUUGAGGUAGCGCACAUACCAUUUGGUGCGCGAUCCGCUCAGGUUUCUUCUGUUUGGAUCACCCUUGCCACCGGGUUGUCCC---GGUGACGUUAUAUUCGGUUCGCGUUUCUGACGC ...((((..((((((..........))))))((((((..((((.....))))..))))))..))))..(((((((....)))---)))).(((((.(..((.....))..).))))). ( -44.90, z-score = -3.27, R) >consensus CUGGAGGUAACGAAGAUACCACUUGGUCCUCGAACCGCUCAGAUUUCUACGGUUAGGGUCAUCCAGGCCACCGGGUUGACCC__UGGUGGCAAUAGAUUUCCAUCCCGUUU__G_CGC .....((...(((.(((........))).)))..))...(((((((..(((....(((((((((........))).)))))).....)))....)))))))................. (-12.99 = -13.23 + 0.25)

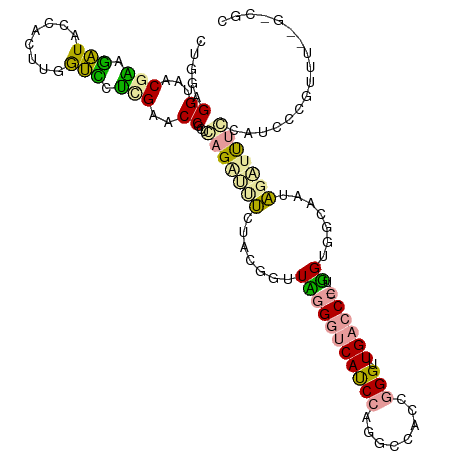
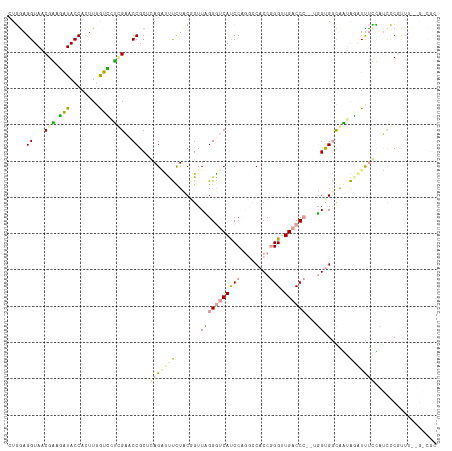
| Location | 183,190 – 183,310 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.89 |
| Shannon entropy | 0.52508 |
| G+C content | 0.57222 |
| Mean single sequence MFE | -48.30 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.13 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 183190 120 + 2517507 GUCAUCCUGGGGCCCGGGAUGACCCUAACCGUAAGGGGCUGAGCGGGGCCAUAACGAAGUGGUAUCUUCGUUACCUUCAGCAGGGCAAGUCACCCGCAGAAGCUUUGUCUCUGGCCAAGA ((((((((((...)))))))))).....((....))(((((((((((((..((((((((......))))))))..(((.((.(((.......))))).))))))))).))).)))).... ( -50.60, z-score = -1.95, R) >droGri2.scaffold_14822 128589 120 - 1097346 GUCAACCCGGUGGGCUGGAUGACCCGAACCGGAGAAAUCUCUGCGGUUCAAGGGUCAAGUGGUACCUCCGGUACCUCCAGCAGGGACUCAACCCCGAUGAAGCCCUGAAAAAGGCCAAAG ........(((..((((((((((((((((((.(((....))).))))))..))))))...((((((...))))))))))))((((..(((.......)))..)))).......))).... ( -52.40, z-score = -3.42, R) >droVir3.scaffold_12823 182002 120 - 2474545 GACAACCCGGUGGCAAGGGUGAUCCAAACAGAAGAAACCUGAGCGGAUCGCGCACCAAAUGGUAUGUGCGCUACCUCAAAAAGGGAUUUCAUCCCGAUGAAGCCUUGCGCAUGGCCAGGG .....(((((((((..(.(((((((...(((.......)))...))))))).)(((....)))......))))))(((....(((((...)))))..))).(((........)))..))) ( -41.90, z-score = -1.61, R) >consensus GUCAACCCGGUGGCCAGGAUGACCCAAACCGAAGAAACCUGAGCGGAUCAAGAACCAAGUGGUAUCUCCGCUACCUCCAGCAGGGAAUCAAACCCGAUGAAGCCUUGAAAAUGGCCAAGG .....(((..(((((.((.....))......(((....)))....)))))..........((((.......)))).......)))................(((........)))..... (-14.68 = -14.13 + -0.54)

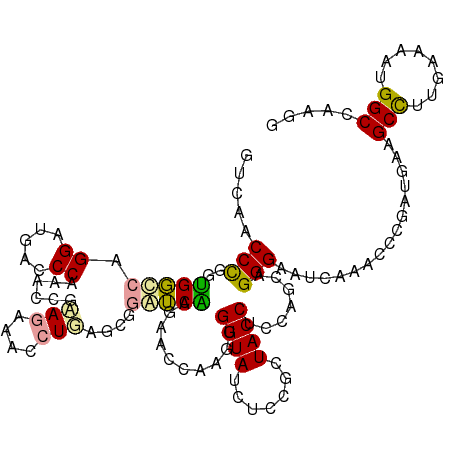
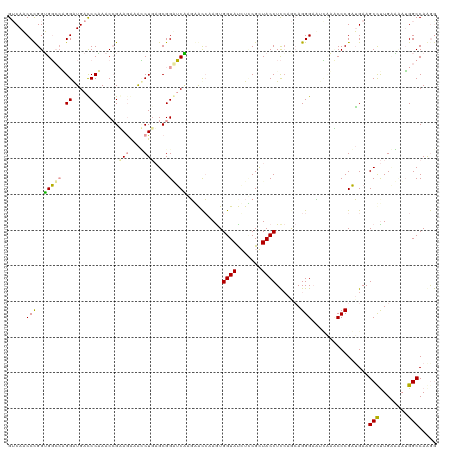
| Location | 183,190 – 183,310 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.89 |
| Shannon entropy | 0.52508 |
| G+C content | 0.57222 |
| Mean single sequence MFE | -49.47 |
| Consensus MFE | -20.75 |
| Energy contribution | -17.33 |
| Covariance contribution | -3.42 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 183190 120 - 2517507 UCUUGGCCAGAGACAAAGCUUCUGCGGGUGACUUGCCCUGCUGAAGGUAACGAAGAUACCACUUCGUUAUGGCCCCGCUCAGCCCCUUACGGUUAGGGUCAUCCCGGGCCCCAGGAUGAC ((((((............((((.((((((.....)))).)).))))(((((((((......)))))))))(((((.....((((......)))).(((....)))))))))))))).... ( -46.30, z-score = -1.80, R) >droGri2.scaffold_14822 128589 120 + 1097346 CUUUGGCCUUUUUCAGGGCUUCAUCGGGGUUGAGUCCCUGCUGGAGGUACCGGAGGUACCACUUGACCCUUGAACCGCAGAGAUUUCUCCGGUUCGGGUCAUCCAGCCCACCGGGUUGAC ..(..((((.....((((.((((.......)))).))))((((((((((((...))))))...((((((..((((((..(((....))))))))))))))))))))).....))))..). ( -53.50, z-score = -2.74, R) >droVir3.scaffold_12823 182002 120 + 2474545 CCCUGGCCAUGCGCAAGGCUUCAUCGGGAUGAAAUCCCUUUUUGAGGUAGCGCACAUACCAUUUGGUGCGCGAUCCGCUCAGGUUUCUUCUGUUUGGAUCACCCUUGCCACCGGGUUGUC .(((((...((.(((((((((((..(((((...)))))....)))))..((((((..........))))))((((((..((((.....))))..))))))..)))))))))))))..... ( -48.60, z-score = -3.30, R) >consensus CCUUGGCCAUACACAAGGCUUCAUCGGGAUAAAAUCCCUGCUGGAGGUAACGAAGAUACCACUUGGUCCUCGAACCGCUCAGAUUUCUACGGUUAGGGUCAUCCAGGCCACCGGGUUGAC .(((((...........((......((((.....)))).......((...(((.(((........))).)))..))))...(((((.........)))))..........)))))..... (-20.75 = -17.33 + -3.42)

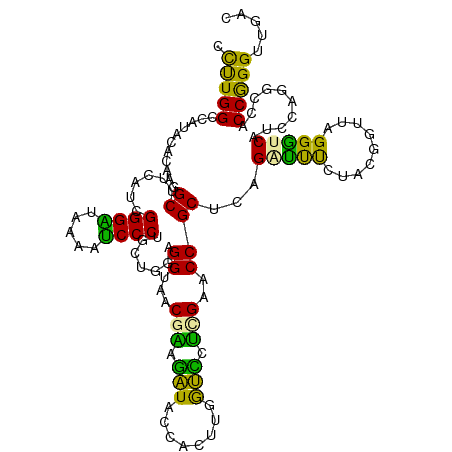
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:33 2011