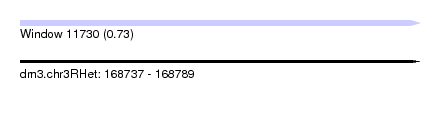
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 168,737 – 168,789 |
| Length | 52 |
| Max. P | 0.730101 |

| Location | 168,737 – 168,789 |
|---|---|
| Length | 52 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 71.91 |
| Shannon entropy | 0.48283 |
| G+C content | 0.40824 |
| Mean single sequence MFE | -13.34 |
| Consensus MFE | -5.08 |
| Energy contribution | -5.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 168737 52 + 2517507 AAUUUUACUGGCGCAGUCGGUAGGAUACA-AAAGUAUCCGAAAAAAAGAACCU---- .....(((((((...)))))))((((((.-...))))))..............---- ( -13.90, z-score = -3.14, R) >droSim1.chr2h_random 1781111 53 + 3178526 CAUUUCACUGGCGCAGUCGGUAGGUUACAUAAAGUAAGCGAAAAAAAUUACCU---- ..((((((((((...)))))).(.((((.....)))).)))))..........---- ( -9.90, z-score = -1.44, R) >droYak2.chrU 14264007 56 + 28119190 AAGUUUAUUGGCGCAGUCGGUAGGAUACUGAAAAAAGUCCUAGUAA-CGCGUUAUCC ........((((((.((.(.((((((..(....)..)))))).).)-)))))))... ( -14.80, z-score = -2.19, R) >droEre2.scaffold_4929 22553542 57 + 26641161 AAGUUUAUUGGCGCAGUCGGUAGGAUACUGAAAAAAGUCCUAGUAAGCUCGUUAUCC .((((((((((.((..((((((...)))))).....)).))))))))))........ ( -13.70, z-score = -1.83, R) >droSec1.super_46 74372 56 + 322281 AAGUUUAUUGGCGCAGUCGGUAGGAUACUGAAAAA-GUUCUAGUAAGCUCGUUAUCC .((((((((((.((..((((((...))))))....-)).))))))))))........ ( -14.40, z-score = -2.27, R) >consensus AAGUUUAUUGGCGCAGUCGGUAGGAUACUGAAAAAAGUCCUAGUAAACUCGUUAUCC ...(((((((((...)))))))))................................. ( -5.08 = -5.04 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:30 2011