
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,218,822 – 6,218,916 |
| Length | 94 |
| Max. P | 0.836479 |

| Location | 6,218,822 – 6,218,916 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.04 |
| Shannon entropy | 0.70584 |
| G+C content | 0.45336 |
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.04 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
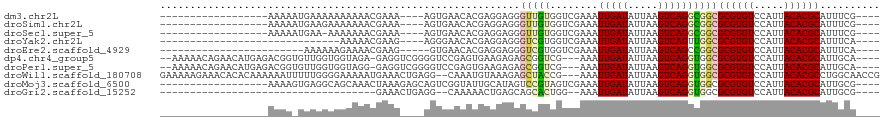
>dm3.chr2L 6218822 94 + 23011544 ------------------AAAAAUGAAAAAAAAAACGAAA----AGUGAACACGAGGAGGGUUGUGGUCGAAAUUGAUAUUAAGUCAGGCGGCGCGUGUCCAUUACACGCAUUUCG---- ------------------.................(((((----......(((((......)))))((((...(((((.....))))).))))((((((.....)))))).)))))---- ( -23.30, z-score = -1.80, R) >droSim1.chr2L 6014183 94 + 22036055 ------------------AAAAAUGAAGAAAAAAACGAAA----AGUGAACACGAGGAGGGUUGUGGUCGAAAUUGAUAUUAAGUCAGGCGGCGCGUGUCCAUUACACGCAUUUCG---- ------------------.................(((((----......(((((......)))))((((...(((((.....))))).))))((((((.....)))))).)))))---- ( -23.30, z-score = -1.70, R) >droSec1.super_5 4290459 93 + 5866729 ------------------AAAAAUGAA-AAAAAAACGAAA----AGUGAACACGAGGAGGGUUGUGGUCGAAAUUGAUAUUAAGUCAGGCGGCGCGUGUCCAUUACACGCAUUUCG---- ------------------.........-.......(((((----......(((((......)))))((((...(((((.....))))).))))((((((.....)))))).)))))---- ( -23.30, z-score = -1.81, R) >droYak2.chr2L 15634320 82 - 22324452 ------------------------------AAAAACGAAG----AGGGAACACGAGGAGGGUCGUGGUCGAAAUUGAUAUUAAGUCAGUUGGCGCGUGUCCAUUACACGCAUUUCA---- ------------------------------......((((----......(((((......)))))(((((...((((.....)))).)))))((((((.....)))))).)))).---- ( -22.10, z-score = -1.62, R) >droEre2.scaffold_4929 15145727 87 + 26641161 ------------------------AAAAAAGAAAACGAAG-----GUGAACACGAGGAGGGUCGUGGUCGAAAUUGAUAUUAAGUCAGCCGGCGCGUGUCCAUUACACGCAUUUCA---- ------------------------......((((..(..(-----((((.(((((......))))).)).....((((.....)))))))..)((((((.....)))))).)))).---- ( -23.20, z-score = -1.33, R) >dp4.chr4_group5 92929 110 - 2436548 --AAAAACAGAACAUGAGACGGUGUUGGUGGUAGA-GAGGUCGGGGUCCGAGUGAAGAGAGCGGUCG---AAAUUGAUAUUAAGUCAGGUGGCGCGUGUCCAUUACACGCAUUGCA---- --...............(((.((((..((....((-..(.((....((.....))...)).)..)).---..))..))))...)))..(..(.((((((.....)))))).)..).---- ( -24.20, z-score = 0.92, R) >droPer1.super_5 92462 110 - 6813705 --AAAAACAGAACAUGAGACGGUGUUGGUGGUAGG-GAGGUCGGGGUCCGAGUGAAGAGAGCGGUCG---AAAUUGAUAUUAAGUCAGGUGGCGCGUGUCCAUUACACGCAUUGCA---- --...............(.((((((..(((....(-(((.((((...)))).).......(((((((---...(((((.....))))).)))).))).)))....)))))))))).---- ( -25.00, z-score = 0.77, R) >droWil1.scaffold_180708 5073440 115 - 12563649 GAAAAAGAAACACACAAAAAAUUUUUGGGGAAAAAUGAAACUGAGG--CAAAUGUAAAGAGCUACCG---AAAUUGAUAUUAAGUCAGGUGGCGCGUGUCCAUUACACGCCUGGCAACCG ..............((((.....))))((...........((...(--(....))..)).((((((.---....((((.....))))))))))((((((.....)))))).......)). ( -21.10, z-score = 0.28, R) >droMoj3.scaffold_6500 19035849 98 - 32352404 ------------------AAAAGUGAGGCAGCAAACUAAAGAGCAGUCGGUAUUGCAUAGUCCGUAGUCGAAAUUGAUAUUAAGUCAGGUGGCGCGUGUCCAUUACACGCAUUGCG---- ------------------............((((........(((((....)))))..........((((...(((((.....))))).))))((((((.....)))))).)))).---- ( -23.20, z-score = -0.25, R) >droGri2.scaffold_15252 14577300 76 - 17193109 ------------------------------------GAAACUGAGG--CAAAAACUGAGCAGCACUGG--AAAUUGAUAUUAAGUCAGGUGGCGCGUGUCCAUUACACGCAUUGCG---- ------------------------------------.........(--(((.......((.((.((((--...(((....))).)))))).))((((((.....)))))).)))).---- ( -19.40, z-score = -0.73, R) >consensus __________________AAAA_UGAAGAAAAAAACGAAA____AGUGAACACGAGGAGGGUCGUGGUCGAAAUUGAUAUUAAGUCAGGUGGCGCGUGUCCAUUACACGCAUUGCA____ ............................................................(((((........(((((.....))))))))))((((((.....)))))).......... (-11.22 = -11.04 + -0.18)

| Location | 6,218,822 – 6,218,916 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.04 |
| Shannon entropy | 0.70584 |
| G+C content | 0.45336 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.53 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6218822 94 - 23011544 ----CGAAAUGCGUGUAAUGGACACGCGCCGCCUGACUUAAUAUCAAUUUCGACCACAACCCUCCUCGUGUUCACU----UUUCGUUUUUUUUUUCAUUUUU------------------ ----(((((.((((((.....))))))........................((.(((..........))).))...----))))).................------------------ ( -14.50, z-score = -0.59, R) >droSim1.chr2L 6014183 94 - 22036055 ----CGAAAUGCGUGUAAUGGACACGCGCCGCCUGACUUAAUAUCAAUUUCGACCACAACCCUCCUCGUGUUCACU----UUUCGUUUUUUUCUUCAUUUUU------------------ ----(((((.((((((.....))))))........................((.(((..........))).))...----))))).................------------------ ( -14.50, z-score = -0.66, R) >droSec1.super_5 4290459 93 - 5866729 ----CGAAAUGCGUGUAAUGGACACGCGCCGCCUGACUUAAUAUCAAUUUCGACCACAACCCUCCUCGUGUUCACU----UUUCGUUUUUUU-UUCAUUUUU------------------ ----(((((.((((((.....))))))........................((.(((..........))).))...----))))).......-.........------------------ ( -14.50, z-score = -0.62, R) >droYak2.chr2L 15634320 82 + 22324452 ----UGAAAUGCGUGUAAUGGACACGCGCCAACUGACUUAAUAUCAAUUUCGACCACGACCCUCCUCGUGUUCCCU----CUUCGUUUUU------------------------------ ----.(((((((((((.....))))))......(((.......))))))))((.(((((......))))).))...----..........------------------------------ ( -17.30, z-score = -2.06, R) >droEre2.scaffold_4929 15145727 87 - 26641161 ----UGAAAUGCGUGUAAUGGACACGCGCCGGCUGACUUAAUAUCAAUUUCGACCACGACCCUCCUCGUGUUCAC-----CUUCGUUUUCUUUUUU------------------------ ----.(((((((((((.....))))))......(((.......))))))))((.(((((......))))).))..-----................------------------------ ( -18.20, z-score = -1.13, R) >dp4.chr4_group5 92929 110 + 2436548 ----UGCAAUGCGUGUAAUGGACACGCGCCACCUGACUUAAUAUCAAUUU---CGACCGCUCUCUUCACUCGGACCCCGACCUC-UCUACCACCAACACCGUCUCAUGUUCUGUUUUU-- ----.(((..(((((..((((....(((.....(((.......)))....---....))).........((((...))))....-.............))))..)))))..)))....-- ( -15.92, z-score = 0.15, R) >droPer1.super_5 92462 110 + 6813705 ----UGCAAUGCGUGUAAUGGACACGCGCCACCUGACUUAAUAUCAAUUU---CGACCGCUCUCUUCACUCGGACCCCGACCUC-CCUACCACCAACACCGUCUCAUGUUCUGUUUUU-- ----.(((..(((((..((((....(((.....(((.......)))....---....))).........((((...))))....-.............))))..)))))..)))....-- ( -15.92, z-score = 0.09, R) >droWil1.scaffold_180708 5073440 115 + 12563649 CGGUUGCCAGGCGUGUAAUGGACACGCGCCACCUGACUUAAUAUCAAUUU---CGGUAGCUCUUUACAUUUG--CCUCAGUUUCAUUUUUCCCCAAAAAUUUUUUGUGUGUUUCUUUUUC .(((((((.(((((((.......)))))))...(((.......)))....---.)))))))..........(--((.(((....((((((....))))))...))).).))......... ( -20.50, z-score = -0.11, R) >droMoj3.scaffold_6500 19035849 98 + 32352404 ----CGCAAUGCGUGUAAUGGACACGCGCCACCUGACUUAAUAUCAAUUUCGACUACGGACUAUGCAAUACCGACUGCUCUUUAGUUUGCUGCCUCACUUUU------------------ ----.(((..((((((.....)))))).............................(((((((.(((........)))....))))))).))).........------------------ ( -18.90, z-score = -0.73, R) >droGri2.scaffold_15252 14577300 76 + 17193109 ----CGCAAUGCGUGUAAUGGACACGCGCCACCUGACUUAAUAUCAAUUU--CCAGUGCUGCUCAGUUUUUG--CCUCAGUUUC------------------------------------ ----.((((.((((((.....))))))((..((((...............--.))).)..)).......)))--).........------------------------------------ ( -14.09, z-score = 0.33, R) >consensus ____CGAAAUGCGUGUAAUGGACACGCGCCACCUGACUUAAUAUCAAUUUCGACCACAACCCUCCUCGUGUUCACU____UUUCGUUUUUUUCCUCA_UUUU__________________ ..........((((((.....))))))............................................................................................. ( -9.00 = -9.00 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:43 2011