| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,496,289 – 24,496,459 |
| Length | 170 |
| Max. P | 0.908502 |

| Location | 24,496,289 – 24,496,459 |
|---|---|
| Length | 170 |
| Sequences | 4 |
| Columns | 170 |
| Reading direction | forward |
| Mean pairwise identity | 52.10 |
| Shannon entropy | 0.79144 |
| G+C content | 0.41431 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -8.85 |
| Energy contribution | -11.85 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
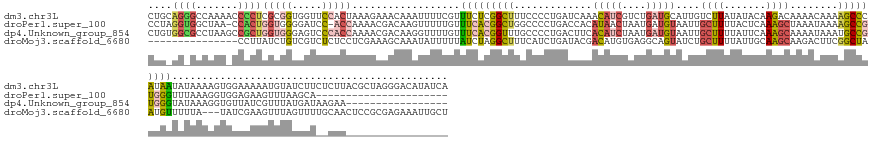
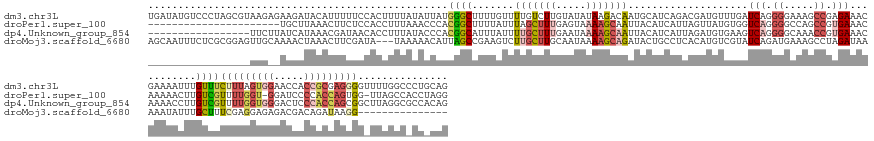
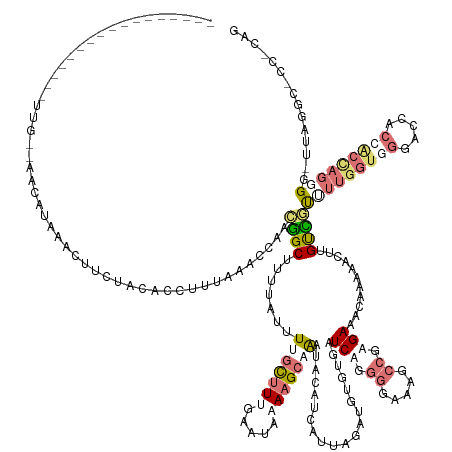
>dm3.chr3L 24496289 170 + 24543557 CUGCAGGGCCAAAACCCCUCGCGGUGGUUCCACUAAAGAAACAAAUUUUCGUUUCUCGGCUUUCCCCUGAUCAAACAUCGUCUGAUGCAUUGUCUUAUAUACAAGACAAAACAAAAGCCCAUAAUAUAAAAGUGGAAAAAUGUAUCUUCUCUUACGCUAGGGACAUAUCA ((((((((.......)))).))))...(((((((..((((((........)))))).((((((............((((....))))..(((((((......)))))))....))))))...........)))))))..........(((((......)))))....... ( -41.80, z-score = -2.77, R) >droPer1.super_100 48496 146 - 203787 CCUAGGUGGCUAA-CCACUGGUGGGGAUCC-ACCAAAACGACAAGUUUUUGUUUCACGGCUGGCCCCUGACCACAUAACUAAUGAUGUAAUUGCUUUUACUCAAAGCUAAAUAAAAGCCGUGGGUUUAAAGGUGGAGAAGUUUAAGCA---------------------- ..((((.(((((.-((..((((((....))-))))((((((.......))))))...)).))))))))).((((.....((((......))))((((.(((((..(((.......)))..)))))..)))))))).............---------------------- ( -41.00, z-score = -0.51, R) >dp4.Unknown_group_854 2336 153 - 3980 CUGUGGCGCCUAAGCCGCUGGUGGGAGUCCCACCAAAACGACAAGGUUUUGUUUCACGGUUUGCCCCUGACUUCACAUCUAAUGAUGUAAUUGCUUUUAUUCAAAGCAAAAUAAAUGCCGUGGGUAUAAAGGUGUUAUCGUUUAUGAUAAGAA----------------- ..(((((......)))))(((((((...)))))))((((((...(.((((((((((((((..............(((((....)))))..(((((((.....))))))).......)))))))).)))))).)....))))))..........----------------- ( -48.40, z-score = -2.79, R) >droMoj3.scaffold_6680 24022688 152 + 24764193 ---------------CCUUAUCUGUCGUCUCUCCUCGAAAGCAAAUAUUUUUAUCUAGGCUUUCAUCUGAUACGACAUGUGAGGCAGUAUCUGCUUUUAUUGCAAGCAAGACUUCGGCUAAUGUUUUUA---UAUCGAAGUUUAGUUUUGCAACUCCGCGAGAAAUUGCU ---------------.(((((.((((((.((.....((((((..(((....)))....))))))....)).)))))).)))))(((((.(((((.....((((((((.(((((((((.(((.....)))---..))))))))).).)))))))....)).))).))))). ( -44.80, z-score = -3.53, R) >consensus CUG_GG_GCCUAA_CCCCUGGCGGGGGUCCCACCAAAAAAACAAAUUUUUGUUUCACGGCUUGCCCCUGACCACACAUCUAAUGAUGUAAUUGCUUUUAUUCAAAGCAAAACAAAAGCCGAGGGUAUAAAGGUGGAAAAGUUUAUGUU__CAA_________________ ....((((.......))))(((((.....)))))..................(((((((((.............(((((....)))))....((((.......))))........))))))))).............................................. ( -8.85 = -11.85 + 3.00)
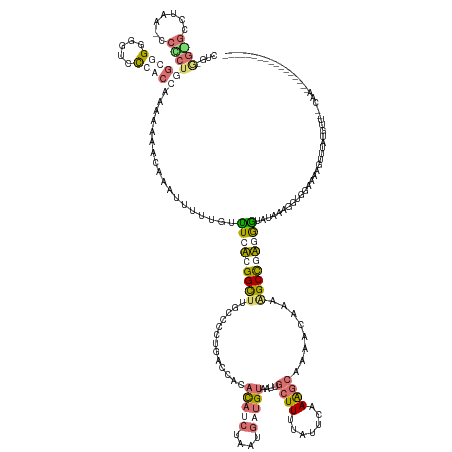
| Location | 24,496,289 – 24,496,459 |
|---|---|
| Length | 170 |
| Sequences | 4 |
| Columns | 170 |
| Reading direction | reverse |
| Mean pairwise identity | 52.10 |
| Shannon entropy | 0.79144 |
| G+C content | 0.41431 |
| Mean single sequence MFE | -43.97 |
| Consensus MFE | -8.10 |
| Energy contribution | -11.60 |
| Covariance contribution | 3.50 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.18 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24496289 170 - 24543557 UGAUAUGUCCCUAGCGUAAGAGAAGAUACAUUUUUCCACUUUUAUAUUAUGGGCUUUUGUUUUGUCUUGUAUAUAAGACAAUGCAUCAGACGAUGUUUGAUCAGGGGAAAGCCGAGAAACGAAAAUUUGUUUCUUUAGUGGAACCACCGCGAGGGGUUUUGGCCCUGCAG ......((((.(((..((((((..((........))..))))))..))).))))...(((.((((((((....)))))))).)))((((((...)))))).(((((.......(((((((((....)))))))))....((((((.((....))))))))..)))))... ( -50.60, z-score = -2.58, R) >droPer1.super_100 48496 146 + 203787 ----------------------UGCUUAAACUUCUCCACCUUUAAACCCACGGCUUUUAUUUAGCUUUGAGUAAAAGCAAUUACAUCAUUAGUUAUGUGGUCAGGGGCCAGCCGUGAAACAAAAACUUGUCGUUUUGGU-GGAUCCCCACCAGUGG-UUAGCCACCUAGG ----------------------................(((......((((((((((((((((....)))))))))))(((((......))))).)))))..((((((.(((((....((((....))))....(((((-((....))))))))))-)).))).)))))) ( -40.50, z-score = -1.32, R) >dp4.Unknown_group_854 2336 153 + 3980 -----------------UUCUUAUCAUAAACGAUAACACCUUUAUACCCACGGCAUUUAUUUUGCUUUGAAUAAAAGCAAUUACAUCAUUAGAUGUGAAGUCAGGGGCAAACCGUGAAACAAAACCUUGUCGUUUUGGUGGGACUCCCACCAGCGGCUUAGGCGCCACAG -----------------...(((((......)))))...............(((.......(((((((.....)))))))(((((((....)))))))..(((.((.....)).))).......(((.(((((..(((((((...))))))))))))..))).))).... ( -46.30, z-score = -3.66, R) >droMoj3.scaffold_6680 24022688 152 - 24764193 AGCAAUUUCUCGCGGAGUUGCAAAACUAAACUUCGAUA---UAAAAACAUUAGCCGAAGUCUUGCUUGCAAUAAAAGCAGAUACUGCCUCACAUGUCGUAUCAGAUGAAAGCCUAGAUAAAAAUAUUUGCUUUCGAGGAGAGACGACAGAUAAGG--------------- .((((((((....))))))))........((((((...---(((.....)))..)))))).((((((.......))))))......(((....((((((.((...(((((((..(((((....))))))))))))..))...))))))....)))--------------- ( -38.50, z-score = -2.09, R) >consensus _________________UUG__AACAUAAACUUCUACACCUUUAAACCAACGGCUUUUAUUUUGCUUUGAAUAAAAGCAAAUACAUCAUUAGAUGUGUGAUCAGGGGAAAGCCGAGAAACAAAAACUUGUCGUUUUGGUGGGACCACCACCAGGGG_UUAGGC_CC_CAG ..................................................((((.......(((((((.....)))))))....................(((.((.....)).)))...........))))(((((((((.....)))))))))............... ( -8.10 = -11.60 + 3.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:21 2011