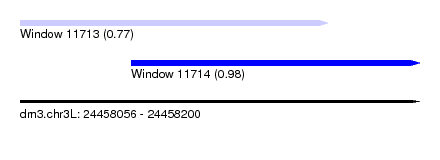
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,458,056 – 24,458,200 |
| Length | 144 |
| Max. P | 0.981264 |

| Location | 24,458,056 – 24,458,167 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.75 |
| Shannon entropy | 0.40537 |
| G+C content | 0.43641 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.77 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.769713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
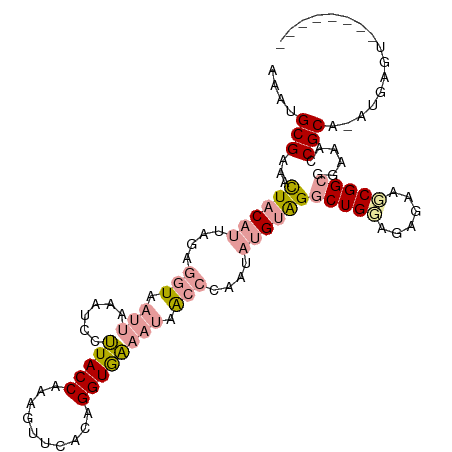
>dm3.chr3L 24458056 111 + 24543557 AGGUGCGACAUUCCCCUUGACAUAAUAAAGUCUGUACCAGAGUUCUCCGGUACCCAAGACGAGUAUGUGGCCUGGAGACAAUCGGCCAUAUACGCCUACGAGCUCUUCAAA .((((((((....................))).)))))(((((((...(((...(.....).((((((((((((....))...)))))))))))))...)))))))..... ( -36.55, z-score = -3.09, R) >droSec1.super_174 27672 102 - 34763 AAAUGCGAAACUACAUUAGAGGUAAUUAAAUCCUUACCAAAGUUCACAGGUGAAAUAACCCAAUAUGUAGGCUGCAGAGAAGCGGCGGAAACCGCA-AUGAGU-------- ...((((...((((((....(((((........)))))..........(((......)))....))))))(((((......)))))(....)))))-......-------- ( -26.50, z-score = -2.67, R) >droSim1.chrU 11322282 102 - 15797150 AAAUGCGAAACUACAUUAGAGGUAAUUAAAUCCUUACCAAAGUUCACAGGUGAAAUAACCCAAUAUGUAGGCUGGAAAGAAGCGGCGGAAACCGCA-AUGAGU-------- ...((((...((((((....(((((........)))))..........(((......)))....))))))((((........))))(....)))))-......-------- ( -22.80, z-score = -1.48, R) >consensus AAAUGCGAAACUACAUUAGAGGUAAUUAAAUCCUUACCAAAGUUCACAGGUGAAAUAACCCAAUAUGUAGGCUGGAGAGAAGCGGCGGAAACCGCA_AUGAGU________ ....(((...((((((....(((.(((......(((((..........)))))))).)))....))))))(((((......)))))......)))................ (-11.98 = -13.77 + 1.79)

| Location | 24,458,096 – 24,458,200 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 52.58 |
| Shannon entropy | 0.75113 |
| G+C content | 0.39125 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -7.45 |
| Energy contribution | -8.07 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
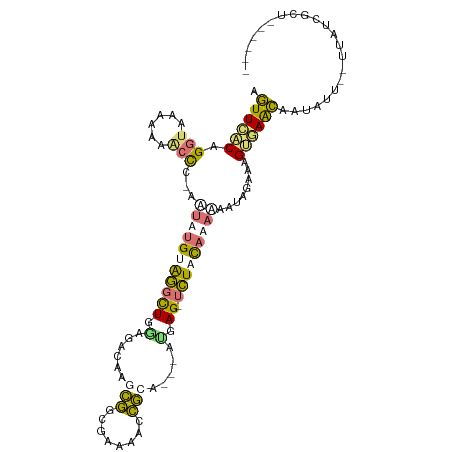
>dm3.chr3L 24458096 104 + 24543557 AGUUCUCCGGUACCCAAGACG-AGUAUGUGGCCUGGAGACAAUCGGCCAUAUACGCCU--ACGAGCUCUUCAAACCAUACAAUGGCAGCAGUGCCCAUUAUCAGGCU---- ......(((((....((((..-.((((((((((((....))...))))))))))((..--....))))))...))).......((((....))))........))..---- ( -29.30, z-score = -0.83, R) >droSec1.super_174 27712 98 - 34763 AGUUCACAGGUGAAAUAACCC-AAUAUGUAGGCUGCAGAGAAGCGGCGGAAACCGCA---AUGAG-UCUAUAUAAAAUUGAAAGUGAACAAUAUU--UUAUCGCU------ .((((((.(((......))).-..((((((((((((......)).((((...)))).---...))-)))))))).........))))))......--........------ ( -28.70, z-score = -3.73, R) >droSim1.chrU 11322322 98 - 15797150 AGUUCACAGGUGAAAUAACCC-AAUAUGUAGGCUGGAAAGAAGCGGCGGAAACCGCA---AUGAG-UCUAUAUAAAAUUGAAAGUGAACAAUAUU--UUAUCGCC------ .((((((.(((......))).-..((((((((((.(......((((......)))).---.).))-)))))))).........))))))......--........------ ( -26.60, z-score = -3.25, R) >triCas2.chrUn_114 8700 111 - 54736 AAUUUACCGGAAAUCAAGCUUUAUUAAGCGAGUUCAUUGCUACAAGUGAACAGUUAAUUAAUAAGUUUUACAAUGUACAAGAUGCAAAUGAUUUUCAAAAUACCUUAUUAA ........((((((((.((((....))))..(((((((......)))))))......................((((.....))))..))))))))............... ( -19.50, z-score = -1.69, R) >consensus AGUUCACAGGUAAAAAAACCC_AAUAUGUAGGCUGGAGACAAGCGGCGAAAACCGCA___AUGAG_UCUACAAAAAAUAGAAAGUGAACAAUAUU__UUAUCGCU______ .((((((.(((......)))...(((((((((..........(((........)))..........)))))))))........))))))...................... ( -7.45 = -8.07 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:17 2011