| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,436,923 – 24,437,013 |
| Length | 90 |
| Max. P | 0.998098 |

| Location | 24,436,923 – 24,437,013 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 58.05 |
| Shannon entropy | 0.54641 |
| G+C content | 0.57932 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -23.29 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
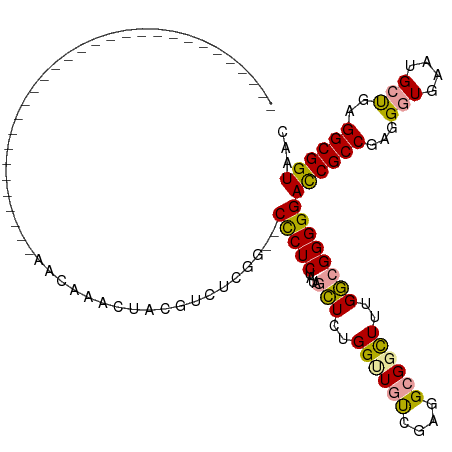
>dm3.chr3L 24436923 90 + 24543557 --------------------------UGCUAACACUCAGCGUUUUG---CCCUCUAGACUUCUGGCUGCCGAGGGGGUUUCGACGGGGGAUCGCCGAGGAUGAAUGGCCAGGCGGUAAC --------------------------........(((..((((..(---((((((.(.(........).).)))))))...)))).)))((((((..((........)).))))))... ( -27.60, z-score = 1.05, R) >droSec1.super_85 23031 84 + 117257 ---------------------------------AUAUUCUGUCUCAG--CUCUCUAUGGCUCUGGUUGUCGAGGCGGCUUUGGCGGGGGACCGCCGAGGGUAAAUGCUGAGGCGGUAAC ---------------------------------.....(((((((((--(((((.(..((....))..).))))...(((((((((....)))))))))......)))))))))).... ( -42.20, z-score = -4.82, R) >droSim1.chr2h_random 1168459 119 - 3178526 AUACUAUGUCUCAGCGUUUAAGUGGGCCGCAACAAAGUACUUCUGGGUGCCCUCUAUAGCUCUGGUUGUCAAGGCGUCUUUGGCGGGGGACCGCCGAGGGUGAAUGCUGAGGCGGUAAC .((((..(((((((((((((((.(((((.((............)).).)))).))...((..((.....))..)).((((((((((....))))))))))))))))))))))))))).. ( -53.70, z-score = -3.84, R) >consensus ______________________________AACAAACUACGUCUCGG__CCCUCUAUAGCUCUGGUUGUCGAGGCGGCUUUGGCGGGGGACCGCCGAGGGUGAAUGCUGAGGCGGUAAC .................................................(((((....(((..((((((....))))))..))))))))((((((...(((....)))..))))))... (-23.29 = -22.97 + -0.32)


| Location | 24,436,923 – 24,437,013 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 58.05 |
| Shannon entropy | 0.54641 |
| G+C content | 0.57932 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24436923 90 - 24543557 GUUACCGCCUGGCCAUUCAUCCUCGGCGAUCCCCCGUCGAAACCCCCUCGGCAGCCAGAAGUCUAGAGGG---CAAAACGCUGAGUGUUAGCA-------------------------- ((((.(((..(((.........((((((......))))))....((((((((........)))..)))))---......)))..))).)))).-------------------------- ( -27.10, z-score = -0.90, R) >droSec1.super_85 23031 84 - 117257 GUUACCGCCUCAGCAUUUACCCUCGGCGGUCCCCCGCCAAAGCCGCCUCGACAACCAGAGCCAUAGAGAG--CUGAGACAGAAUAU--------------------------------- ......(.((((((..........(((((....)))))......(.(((........))).).......)--))))).).......--------------------------------- ( -23.50, z-score = -3.03, R) >droSim1.chr2h_random 1168459 119 + 3178526 GUUACCGCCUCAGCAUUCACCCUCGGCGGUCCCCCGCCAAAGACGCCUUGACAACCAGAGCUAUAGAGGGCACCCAGAAGUACUUUGUUGCGGCCCACUUAAACGCUGAGACAUAGUAU ......((....)).....((((((((((....)))))......((.(((.....))).))....))))).........(((((.(((..((((..........))))..))).))))) ( -33.50, z-score = -1.57, R) >consensus GUUACCGCCUCAGCAUUCACCCUCGGCGGUCCCCCGCCAAAGCCGCCUCGACAACCAGAGCUAUAGAGGG__CCAAGACGCAAUAUGUU______________________________ ...................(((((((((......))))........(((........))).....)))))................................................. (-13.09 = -13.10 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:15 2011