| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,417,925 – 24,418,015 |
| Length | 90 |
| Max. P | 0.665430 |

| Location | 24,417,925 – 24,418,015 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 49.34 |
| Shannon entropy | 0.82241 |
| G+C content | 0.45554 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -6.31 |
| Energy contribution | -6.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
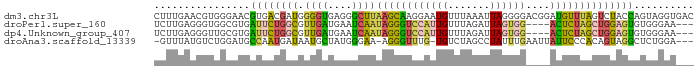
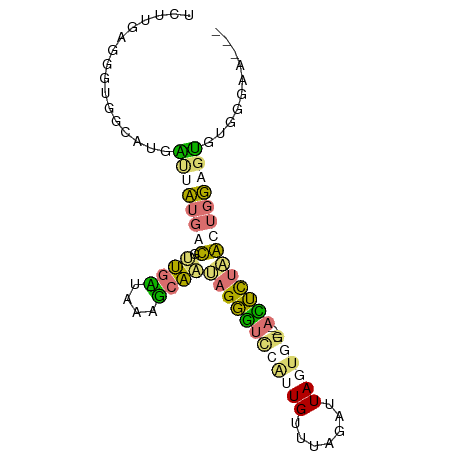
>dm3.chr3L 24417925 90 - 24543557 CUUUGAACGUGGGAACGUGACGAUGGGGUGAGGGCUUAAGCAAGGAAUGUUUAAAUUAGGGGACGGAUGUUUAGUCUACCAGUAGGUGAC ...(((((((..((((((..(....((((....))))......)..))))))......(....)..)))))))(((.(((....)))))) ( -17.40, z-score = -0.15, R) >droPer1.super_160 64127 83 + 79520 UCUUGAGGGUGGCGUGAUUCUGGCGUUGAUGAAUCAAUAGGGUCCAUUGUUUAGAUUAGUGG----ACUCUAGCUGGAGUGUGGGAA--- ................((((..(((((((....)))))(((((((((((.......))))))----))))).))..)))).......--- ( -26.20, z-score = -2.49, R) >dp4.Unknown_group_407 13808 83 - 16403 UCUUGAGGGUUGCGUGAUUCUGGCGUUGAUGAAUCAAUAGGGUCCAUUGUUUAGAUUAGUGG----ACUCUAGCUGGAGUGUGGGAA--- .........(..(...((((..(((((((....)))))(((((((((((.......))))))----))))).))..)))))..)...--- ( -26.30, z-score = -2.65, R) >droAna3.scaffold_13339 7686 84 + 4599533 -GUUUAUGUCUGGAUGCCAAUGAUAAUGCUAUGGGAA-AGGGUUUG-UGUCUAGCCUAUUUGAAUUAUUCCCACAGUAGGCUCUGGA--- -.......((..((.(((........((((.((((((-.(((((..-.....)))))..........)))))).)))))))))..))--- ( -20.80, z-score = -0.27, R) >consensus UCUUGAGGGUGGCAUGAUUAUGACGUUGAUAAAGCAAUAGGGUCCAUUGUUUAGAUUAGUGG____ACUCUAACUGGAGUGUGGGAA___ ................((((((((.((((....))))((((((((((((.......))))))....)))))))))))))).......... ( -6.31 = -6.38 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:12 2011