| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,364,141 – 24,364,243 |
| Length | 102 |
| Max. P | 0.939340 |

| Location | 24,364,141 – 24,364,243 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 57.48 |
| Shannon entropy | 0.78755 |
| G+C content | 0.39445 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -4.18 |
| Energy contribution | -4.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24364141 102 + 24543557 GAUAUCUCUG----UGGAACGCAAAUGGCGUUUCACGGCAUACACAAGAGCUCACACAGUUCAUUUACGAAAAAAACAUCGACGUAAUGCUACUAUCAGAAACGCA ((((.....(----((((((((.....)))))))))(((((((....(((((.....))))).....(((........)))..)).)))))..))))......... ( -27.00, z-score = -3.37, R) >droSim1.chr2h_random 2465968 106 + 3178526 UACGCCUCGGAACCUGGAAAGCUCGUAGCAUUAUCCCCAAUGCUAAUGAGCUUAAGGUGUUUUUUAUCACACACGAAAUUGAUGUUAUGCUUAUCUCAGAGGCAUU ...(((((.((((((...((((((((((((((......)))))).)))))))).))))((....(((((..........)))))....)).....)).)))))... ( -33.50, z-score = -3.73, R) >droSec1.super_67 72388 105 + 170185 UACGCCUCGGAACCUGGAA-GCUCGUAGCAUUAUCCCCAAUGCUAAUGAGCUUAAAGUGUUUCUUAGCACACACGAAAUUGAUGUUAUGCUUAUCUCAGAGGCAUU ...((((((((((((..((-((((((((((((......)))))).))))))))..)).)))))..((((.(((((....)).)))..)))).......)))))... ( -36.70, z-score = -4.69, R) >droVir3.scaffold_13036 142243 97 - 1135500 ----GAUCGGAUUAUGGAACGCGGAUGGUCUUUAUAUUAAAAUC--UGAACUGACC---UUCCUUACCACCCACAACAGCGACAUUAUGCUCCUAACGGAAACUUA ----....((....(((.....(((.((((..............--......))))---.)))...))).))......(((......)))(((....)))...... ( -15.15, z-score = -0.26, R) >anoGam1.chr3R 11775561 102 - 53272125 --AGUUUCCUA--CUGGAAUGCACAUAGCAUUUCAAAGAAAAAACUAGACCUAAUUAAUUUUCUCAACUCAAAUGAAAUAGAUAUUAUGGCAAUAACAGAAACAUU --.(((((...--.((((((((.....))))))))............(.((((((....((((...........)))).....)))).))).......)))))... ( -16.50, z-score = -2.05, R) >consensus _ACGCCUCGGA__CUGGAACGCACAUAGCAUUUCACCCAAUACAAAUGAGCUUAAAGAGUUUCUUAACACACACGAAAUCGAUGUUAUGCUAAUAUCAGAAACAUU ...(((((............((.....))..................((((.......))))....................................)))))... ( -4.18 = -4.78 + 0.60)

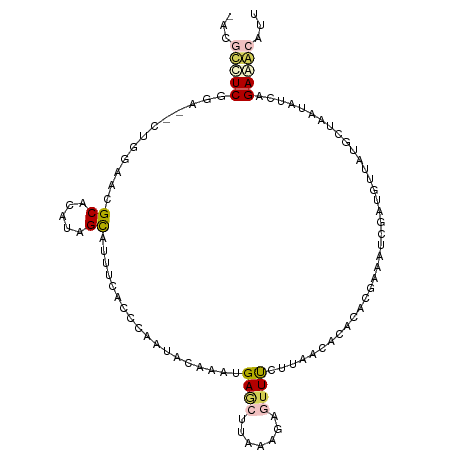
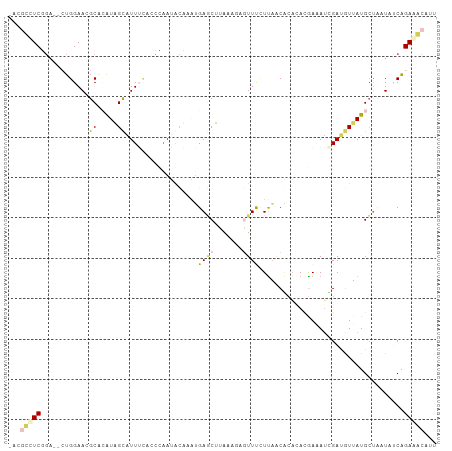
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:10 2011