| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,217,636 – 6,217,739 |
| Length | 103 |
| Max. P | 0.500000 |

| Location | 6,217,636 – 6,217,739 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
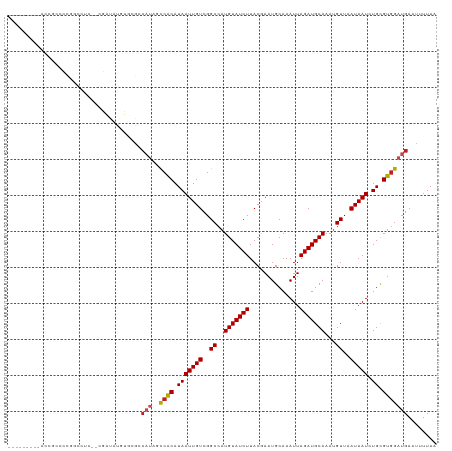
| Mean pairwise identity | 78.58 |
| Shannon entropy | 0.46912 |
| G+C content | 0.40223 |
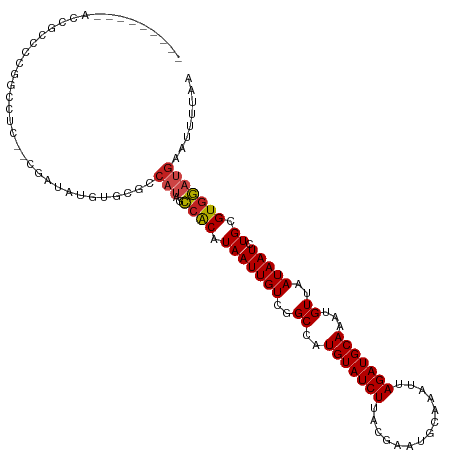
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.57 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 6217636 103 - 23011544 --------------UCCGCCAUC--CGAUACGUUCGCCAUAGCCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA --------------.....((((--((((((((........((((((....)).)))).)))))))......(((((.((((...((....))...)))).))))))))))........ ( -26.40, z-score = -1.57, R) >droSim1.chr2L 6012971 108 - 22036055 ---------AACGCCCCUGCCUC--CGAUACGUUCGCCAUAGCCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ---------..............--.....((((((((((.((((((....)).)))).)))............(((.((((...((....))...)))).))))))))))........ ( -22.80, z-score = -0.26, R) >droSec1.super_5 4289185 108 - 5866729 ---------AACGCCCCUGCCUC--CGAUACGUUCGCCAUAGCCGCAUAAUUGUCGGCCUUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ---------..............--.....(((((((((..((((((....)).)))).((((((((..............))))))))............)).)))))))........ ( -22.04, z-score = -0.19, R) >droYak2.chr2L 15633156 108 + 22324452 ---------ACCGCCCUUGCCUG--CGAUUUGUUCGCCAUAGCCACAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ---------..(((........)--))(((..((((((((.((((((....))).))).)))............(((.((((...((....))...)))).))))))))..)))..... ( -23.80, z-score = -0.47, R) >droEre2.scaffold_4929 15144589 92 - 26641161 ---------------------------AUUUGUUCGCCAUAGCCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ---------------------------(((..((((((((.((((((....)).)))).)))............(((.((((...((....))...)))).))))))))..)))..... ( -22.50, z-score = -1.34, R) >droAna3.scaffold_12916 8192504 100 + 16180835 ---------AUCCCCCCGGUCAC--CGAUAUAUG--------CCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ---------..........((((--(((((((((--------(((((....)).))).))))))))......(((((.((((...((....))...)))).))))))).)))....... ( -25.70, z-score = -1.85, R) >dp4.chr4_group5 91616 118 + 2436548 CACACCCCUACUGCAGCAGGCCCU-UCCCUUCGGAGGCAUAGCCACAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA .......((((.((((..((((..-(((....)))(((...)))...........)))).(((((((..............)))))))............))))))))........... ( -27.34, z-score = -0.36, R) >droPer1.super_5 91122 118 + 6813705 CACACCCCUACUGGUGCAGGCCCUCCCCCUUCGGAGGCAUAGCCACAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGUGU-GAUGAAUUUUAA ..((((......))))..((((((((......)))))....)))((((((((((..((..(((((((..............)))))))...))..))))).)))))-............ ( -29.34, z-score = -1.04, R) >droWil1.scaffold_180708 5071504 82 + 12563649 -------------------------------------CCGAUCCGCAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA -------------------------------------...((((((.(((((((..((..(((((((..............)))))))...))..))))).)).))))))......... ( -17.84, z-score = -1.21, R) >droVir3.scaffold_12963 3013959 108 - 20206255 ------CUCUCUGCUCCG-CAUU--CAAUAUGUGCGGCAU--CCACAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ------.........(((-(((.--......))))))(((--((((.(((((((..((..(((((((..............)))))))...))..))))).)).)))))))........ ( -29.04, z-score = -2.29, R) >droMoj3.scaffold_6500 13322269 111 - 32352404 ----CUCCUACCGCCGCCUGGCA--AUAUGUGUGCGGCAU--CCACAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ----......(((((((.((...--.)).))).))))(((--((((.(((((((..((..(((((((..............)))))))...))..))))).)).)))))))........ ( -29.64, z-score = -1.39, R) >droGri2.scaffold_15252 2620124 108 - 17193109 -------CUCCAGCCUCCUGCUU--CGAUAUGUGCGGCAU--UCACAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGAAUGAAUUUUAA -------..........((((..--........))))(((--((((.(((((((..((..(((((((..............)))))))...))..))))).)).)))))))........ ( -21.74, z-score = -0.30, R) >consensus _________ACCGCCCCGGCCUC__CGAUAUGUGCGCCAUAGCCACAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAA ..........................................((((.(((((((..((..(((((((..............)))))))...))..))))).)).))))........... (-14.89 = -14.57 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:42 2011