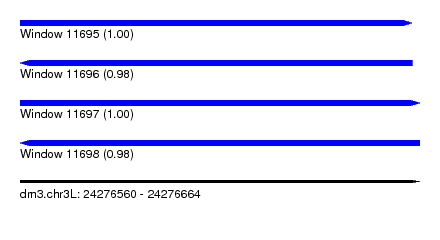
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,276,560 – 24,276,664 |
| Length | 104 |
| Max. P | 0.999868 |

| Location | 24,276,560 – 24,276,662 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 61.95 |
| Shannon entropy | 0.58658 |
| G+C content | 0.55172 |
| Mean single sequence MFE | -34.91 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24276560 102 + 24543557 ------GCACCGAAGAUCUAC-ACACAAUUAAUUUGUGUCACUCACUUCUUCUCCGUGCUAUCCUU--------CGAACCACACUCGGCGACAGCGACCACACGCUGCCGCCGCGCU ------((((.(((((.....-((((((.....))))))........)))))...)))).......--------.........(.(((((.(((((......))))).))))).).. ( -31.72, z-score = -5.05, R) >droYak2.chrU 20653638 109 - 28119190 -------GACUGAAGUUCUAC-GCACAACUGCCGUGUGUCGCACACCUCUUCUACGUGCUUACCACAUGUGCAGCGAAACACACUCGGCAUCAGCAACAACACGCUGGCGCCGCGCU -------.............(-((....((((((((((..((((...........))))....)))))).))))............(((..((((........))))..)))))).. ( -33.70, z-score = -1.12, R) >droAna3.scaffold_13260 592172 109 + 2125536 UGUACACUGUUCCAGUUCUGGUACGCAACUGCCGGCGUCCAACUGUGUACCUACUGUGCAUACUAC--------CAAACCUCGAUCGGCGACAGCGACACAACGCUGUCGCCUCGCU ((((((..((.(((((..(((.((((........))))))))))).).))....))))))......--------.......(((..((((((((((......))))))))))))).. ( -38.70, z-score = -3.06, R) >droMoj3.scaffold_6498 3063543 102 + 3408170 ------GCACUUGAAUUCUGG-ACGCAACUAACUUGUGUCUCUCAAUUAUUCUUUGUGCCUACCAU--------CGAACCUCACUCGGCGACAGCGACACGACGCUGUCGCCCCGCU ------((((..((((...((-((((((.....)))))))).......))))...)))).......--------............((((((((((......))))))))))..... ( -35.50, z-score = -5.60, R) >consensus ______GCACUGAAGUUCUAC_ACACAACUAACGUGUGUCACUCACUUAUUCUCCGUGCUUACCAC________CGAACCACACUCGGCGACAGCGACAACACGCUGUCGCCGCGCU ......................................................................................((((((((((......))))))))))..... (-17.15 = -17.65 + 0.50)


| Location | 24,276,560 – 24,276,662 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 61.95 |
| Shannon entropy | 0.58658 |
| G+C content | 0.55172 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24276560 102 - 24543557 AGCGCGGCGGCAGCGUGUGGUCGCUGUCGCCGAGUGUGGUUCG--------AAGGAUAGCACGGAGAAGAAGUGAGUGACACAAAUUAAUUGUGU-GUAGAUCUUCGGUGC------ .((((((((((((((......))))))))))..((((...((.--------...))..))))...(((((..((....((((((.....))))))-.))..))))).))))------ ( -42.30, z-score = -3.81, R) >droYak2.chrU 20653638 109 + 28119190 AGCGCGGCGCCAGCGUGUUGUUGCUGAUGCCGAGUGUGUUUCGCUGCACAUGUGGUAAGCACGUAGAAGAGGUGUGCGACACACGGCAGUUGUGC-GUAGAACUUCAGUC------- .((((((((((...((((.(((((...(((((.((((((......)))))).))))).((((.(.....).)))))))))))))))).)))))))-..............------- ( -41.50, z-score = -0.32, R) >droAna3.scaffold_13260 592172 109 - 2125536 AGCGAGGCGACAGCGUUGUGUCGCUGUCGCCGAUCGAGGUUUG--------GUAGUAUGCACAGUAGGUACACAGUUGGACGCCGGCAGUUGCGUACCAGAACUGGAACAGUGUACA ..(((((((((((((......))))))))))..))).......--------...((((((.((((.(((((.(((((((...)))))...)).)))))...)))).....)))))). ( -41.50, z-score = -1.45, R) >droMoj3.scaffold_6498 3063543 102 - 3408170 AGCGGGGCGACAGCGUCGUGUCGCUGUCGCCGAGUGAGGUUCG--------AUGGUAGGCACAAAGAAUAAUUGAGAGACACAAGUUAGUUGCGU-CCAGAAUUCAAGUGC------ .(((.((((((((((......))))))))))((((...((((.--------.((.......))..))))......(.(((.(((.....))).))-))...))))...)))------ ( -31.60, z-score = -1.09, R) >consensus AGCGCGGCGACAGCGUGGUGUCGCUGUCGCCGAGUGAGGUUCG________AUGGUAAGCACAGAGAAGAAGUGAGUGACACAAGGCAGUUGCGU_CCAGAACUUCAGUGC______ .((((((((((((((......))))))))))..((((.(((((...........(....)............))))).)))).........))))...................... (-21.67 = -22.43 + 0.75)

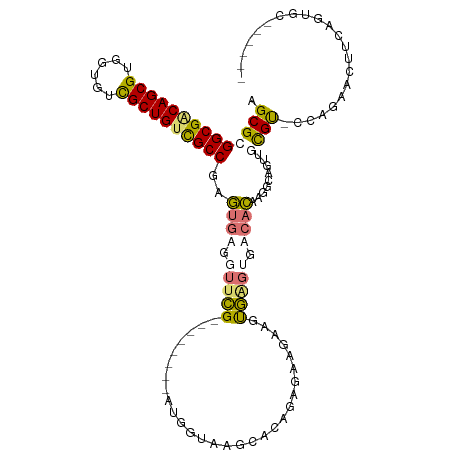
| Location | 24,276,560 – 24,276,664 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 61.99 |
| Shannon entropy | 0.59247 |
| G+C content | 0.54637 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24276560 104 + 24543557 ----GCACCGAAGAUCUAC-ACACAAUUAAUUUGUGUCACUCACUUCUUCUCCGUGCUAUCCUU--------CGAACCACACUCGGCGACAGCGACCACACGCUGCCGCCGCGCUGU ----((((.(((((.....-((((((.....))))))........)))))...)))).......--------......((((.(((((.(((((......))))).))))).).))) ( -33.32, z-score = -4.86, R) >droYak2.chrU 20653638 111 - 28119190 -----GACUGAAGUUCUAC-GCACAACUGCCGUGUGUCGCACACCUCUUCUACGUGCUUACCACAUGUGCAGCGAAACACACUCGGCAUCAGCAACAACACGCUGGCGCCGCGCUAU -----.............(-((....((((((((((..((((...........))))....)))))).))))............(((..((((........))))..)))))).... ( -33.70, z-score = -1.12, R) >droAna3.scaffold_13260 592174 109 + 2125536 UACACUGUUCCAGUUCUGGUACGCAACUGCCGGCGUCCAACUGUGUACCUACUGUGCAUACUAC--------CAAACCUCGAUCGGCGACAGCGACACAACGCUGUCGCCUCGCUAC .....(((..((((...((((((((..((........))..)))))))).)))).)))......--------.......(((..((((((((((......))))))))))))).... ( -37.80, z-score = -3.29, R) >droMoj3.scaffold_6498 3063543 104 + 3408170 ----GCACUUGAAUUCUGG-ACGCAACUAACUUGUGUCUCUCAAUUAUUCUUUGUGCCUACCAU--------CGAACCUCACUCGGCGACAGCGACACGACGCUGUCGCCCCGCUAA ----((((..((((...((-((((((.....)))))))).......))))...)))).......--------............((((((((((......))))))))))....... ( -35.50, z-score = -5.67, R) >consensus ____GCACUGAAGUUCUAC_ACACAACUAACGUGUGUCACUCACUUAUUCUCCGUGCUUACCAC________CGAACCACACUCGGCGACAGCGACAACACGCUGUCGCCGCGCUAU ....................................................................................((((((((((......))))))))))....... (-17.15 = -17.65 + 0.50)


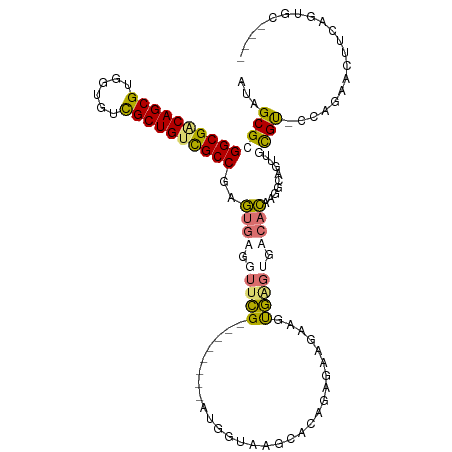
| Location | 24,276,560 – 24,276,664 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 61.99 |
| Shannon entropy | 0.59247 |
| G+C content | 0.54637 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24276560 104 - 24543557 ACAGCGCGGCGGCAGCGUGUGGUCGCUGUCGCCGAGUGUGGUUCG--------AAGGAUAGCACGGAGAAGAAGUGAGUGACACAAAUUAAUUGUGU-GUAGAUCUUCGGUGC---- ...((((((((((((((......))))))))))..((((...((.--------...))..))))...(((((..((....((((((.....))))))-.))..))))).))))---- ( -42.30, z-score = -3.38, R) >droYak2.chrU 20653638 111 + 28119190 AUAGCGCGGCGCCAGCGUGUUGUUGCUGAUGCCGAGUGUGUUUCGCUGCACAUGUGGUAAGCACGUAGAAGAGGUGUGCGACACACGGCAGUUGUGC-GUAGAACUUCAGUC----- ...((((((((((...((((.(((((...(((((.((((((......)))))).))))).((((.(.....).)))))))))))))))).)))))))-..............----- ( -41.50, z-score = -0.17, R) >droAna3.scaffold_13260 592174 109 - 2125536 GUAGCGAGGCGACAGCGUUGUGUCGCUGUCGCCGAUCGAGGUUUG--------GUAGUAUGCACAGUAGGUACACAGUUGGACGCCGGCAGUUGCGUACCAGAACUGGAACAGUGUA (((((..((((((((((......))))))))))(.(((..(((..--------(..((.(((.......))).))..)..)))..)))).)))))((.(((....))).))...... ( -40.50, z-score = -1.30, R) >droMoj3.scaffold_6498 3063543 104 - 3408170 UUAGCGGGGCGACAGCGUCGUGUCGCUGUCGCCGAGUGAGGUUCG--------AUGGUAGGCACAAAGAAUAAUUGAGAGACACAAGUUAGUUGCGU-CCAGAAUUCAAGUGC---- ...(((.((((((((((......))))))))))((((...((((.--------.((.......))..))))......(.(((.(((.....))).))-))...))))...)))---- ( -31.60, z-score = -1.14, R) >consensus AUAGCGCGGCGACAGCGUGGUGUCGCUGUCGCCGAGUGAGGUUCG________AUGGUAAGCACAGAGAAGAAGUGAGUGACACAAGGCAGUUGCGU_CCAGAACUUCAGUGC____ (((((..((((((((((......))))))))))..((((.(((((...........(....)............))))).))))......)))))...................... (-22.10 = -22.85 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:05 2011