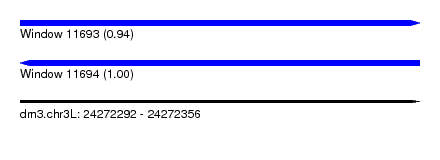
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,272,292 – 24,272,356 |
| Length | 64 |
| Max. P | 0.998935 |

| Location | 24,272,292 – 24,272,356 |
|---|---|
| Length | 64 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 57.55 |
| Shannon entropy | 0.77657 |
| G+C content | 0.50194 |
| Mean single sequence MFE | -14.73 |
| Consensus MFE | -7.64 |
| Energy contribution | -7.17 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
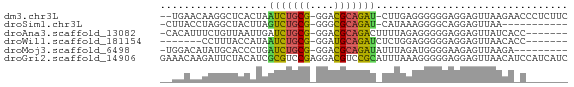
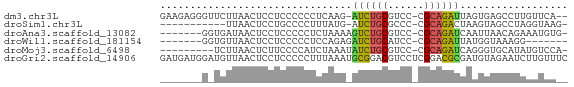
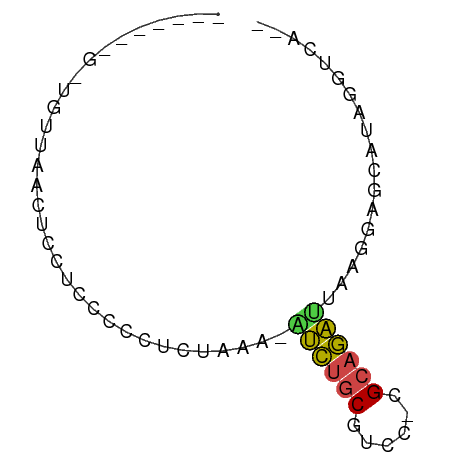
>dm3.chr3L 24272292 64 + 24543557 --UGAACAAGGCUCACUAAUCUGCG-GGACGCAGAU-CUUGAGGGGGGAGGAGUUAAGAACCCUCUUC --.........((((...(((((((-...)))))))-..)))).(((((((.((.....))))))))) ( -20.50, z-score = -1.32, R) >droSim1.chr3L 2988948 54 - 22553184 -CUUACCUAGGCUACUUAGUCUGCG-GGGCGCAGAU-CAUAAAGGGGCAGGAGUUAA----------- -....(((..((..(((.(((((((-...)))))))-....)))..)))))......----------- ( -15.10, z-score = -0.84, R) >droAna3.scaffold_13082 1758569 59 - 3919855 -CACAUUUCUGUUAAUUGAUCUGCG-GGACGCAGACUUUUAGAGGGGGAGGAGUUAUCACC------- -..(.(..((.((((..(.((((((-...)))))))..)))).))..).)...........------- ( -12.40, z-score = -0.56, R) >droWil1.scaffold_181154 4133291 53 + 4610121 -------CCUUUACCAUAAUCUGCG-GGAUGCAGAUCUCUGGAGGGGGAGGAGUUAACACC------- -------((((..(((..((((((.-....))))))...)))..)))).............------- ( -15.00, z-score = -0.70, R) >droMoj3.scaffold_6498 3059478 57 + 3408170 -UGGACAUAUGCACCCUGAUCUGCG-GGACGCAGAUAUUUAGAUGGGGAAGAGUUAAGA--------- -............((((.(((((((-...)))))))........))))...........--------- ( -12.40, z-score = -0.95, R) >droGri2.scaffold_14906 13489288 68 + 14172833 GAAACAAGAUUCUACAUCGCGUCCGAGGACGUCCGCAUUUAAAGGGGGAGGAGUUAACAUCCAUCAUC .......((((((.(...(((((....)))))((.(.......).))).))))))............. ( -13.00, z-score = 0.06, R) >consensus __AGAAAUAUGCUACAUAAUCUGCG_GGACGCAGAU_UUUAAAGGGGGAGGAGUUAACA_C_______ ..................(((((((....)))))))................................ ( -7.64 = -7.17 + -0.47)
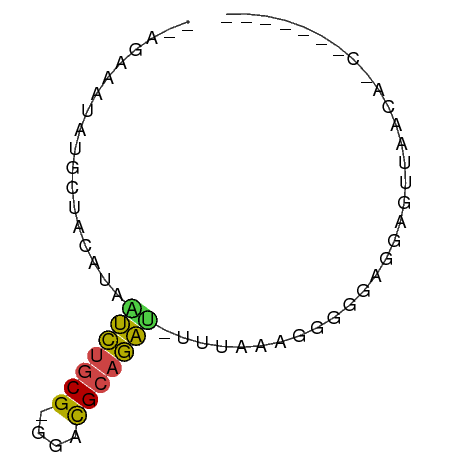
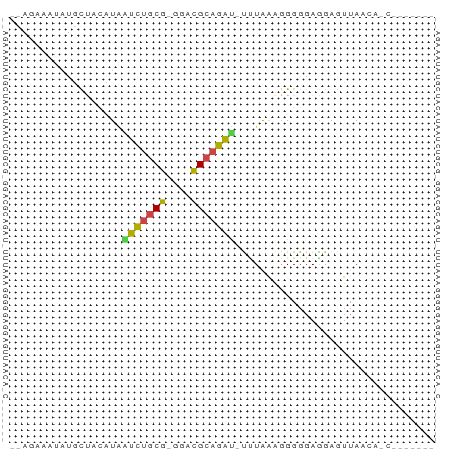
| Location | 24,272,292 – 24,272,356 |
|---|---|
| Length | 64 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 57.55 |
| Shannon entropy | 0.77657 |
| G+C content | 0.50194 |
| Mean single sequence MFE | -13.50 |
| Consensus MFE | -7.45 |
| Energy contribution | -7.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24272292 64 - 24543557 GAAGAGGGUUCUUAACUCCUCCCCCCUCAAG-AUCUGCGUCC-CGCAGAUUAGUGAGCCUUGUUCA-- (((.(((((((...(((.............(-(((((((...-)))))))))))))))))).))).-- ( -20.41, z-score = -2.96, R) >droSim1.chr3L 2988948 54 + 22553184 -----------UUAACUCCUGCCCCUUUAUG-AUCUGCGCCC-CGCAGACUAAGUAGCCUAGGUAAG- -----------......(((((..(((...(-.((((((...-))))))).)))..))..)))....- ( -12.30, z-score = -1.81, R) >droAna3.scaffold_13082 1758569 59 + 3919855 -------GGUGAUAACUCCUCCCCCUCUAAAAGUCUGCGUCC-CGCAGAUCAAUUAACAGAAAUGUG- -------((........)).............(((((((...-)))))))......(((....))).- ( -10.70, z-score = -2.47, R) >droWil1.scaffold_181154 4133291 53 - 4610121 -------GGUGUUAACUCCUCCCCCUCCAGAGAUCUGCAUCC-CGCAGAUUAUGGUAAAGG------- -------((.(....).))....((((((..(((((((....-.))))))).)))...)))------- ( -14.30, z-score = -1.85, R) >droMoj3.scaffold_6498 3059478 57 - 3408170 ---------UCUUAACUCUUCCCCAUCUAAAUAUCUGCGUCC-CGCAGAUCAGGGUGCAUAUGUCCA- ---------..............(((((....(((((((...-)))))))..)))))..........- ( -11.20, z-score = -1.90, R) >droGri2.scaffold_14906 13489288 68 - 14172833 GAUGAUGGAUGUUAACUCCUCCCCCUUUAAAUGCGGACGUCCUCGGACGCGAUGUAGAAUCUUGUUUC ...((.(((((((..(................)..)))))))))(((((.(((.....))).))))). ( -12.09, z-score = 0.43, R) >consensus _______G_UGUUAACUCCUCCCCCUCUAAA_AUCUGCGUCC_CGCAGAUUAAGGAGCAUAGGUCA__ ................................(((((((....))))))).................. ( -7.45 = -7.28 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:47:02 2011