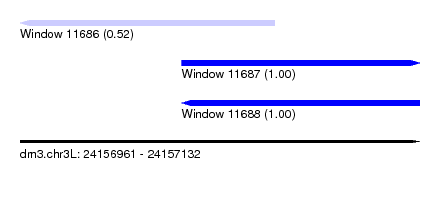
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,156,961 – 24,157,132 |
| Length | 171 |
| Max. P | 0.998112 |

| Location | 24,156,961 – 24,157,070 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 59.23 |
| Shannon entropy | 0.67716 |
| G+C content | 0.54661 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -14.24 |
| Energy contribution | -17.30 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
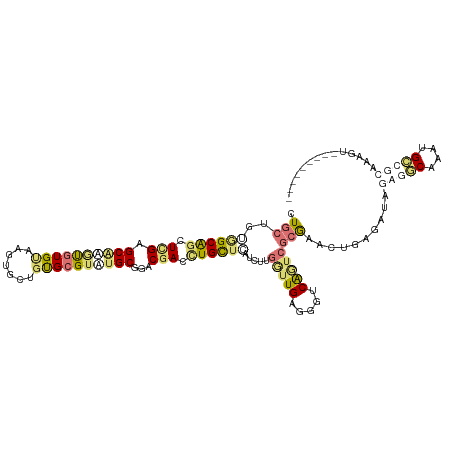
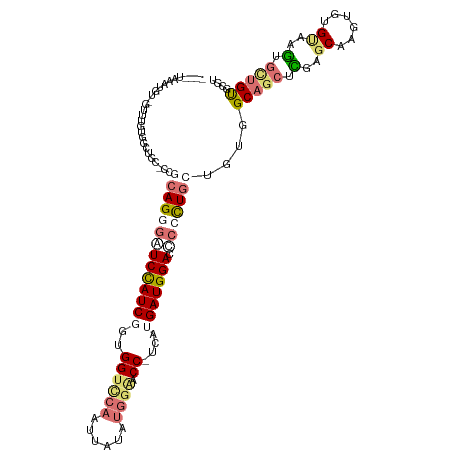
>dm3.chr3L 24156961 109 - 24543557 CUUCUCCAGCGUCUGGAGCCACUGUGUGGUUUCAGCGCGUUUGCAGACGACUUGUUGCUUUUCGUUGAGGAUCGUACCCAAAUCGUAAUGCAGUCGAAAGGCUAACAGC--------- ........(((.((((((((((...)))))))))))))(((.(((.((((.(((.(((..(((.....)))..)))..))).))))..)))((((....)))))))...--------- ( -37.00, z-score = -1.76, R) >droAna3.scaffold_9904 1421 107 + 1642 CUGCUGUGGCAGCUCGAGCAAGCGUGGCAGUGUUGUGCGUAUGCGGACGACUUGCUCAUCUUGGUUGAGGGCCAGUCGCGUUCUAUGAUUGAGGCAAGCGCCUCAGA----------- ..((..((((..(((((.((((.(((((((.(((((.((....)).))))))))).))))))).))))).))))...)).........(((((((....))))))).----------- ( -46.00, z-score = -1.59, R) >droVir3.scaffold_9533 3752 118 + 3988 UUGCUGGUGCAGCUUGAGCAGAUGUGUAAAUGUUGUGCGUAUGCGGACGAUCUGCUUAUCUUGAUUGAGGGUCGGUCGCGUCUUGAGAUCGAGACGAAUGCGGGUAUGUGCUUGCGCA ..((.((..(((((((.(((((((.(((...(((((.((....)).))))).))).))))).(((((.....)))))))((((((....)))))).....))))).))..)).))... ( -38.70, z-score = -0.17, R) >droGri2.scaffold_15056 4768 118 - 38174 UUGCUAUGGCAGCUCGAGCGUGUAUGCAAGUGCUGUGCGUAUGCGGACGACCUGCUCAUUCUGGUUGAGGGGCAAUCGCGAGCGGAUGUAGAGGCAAGUGCGGCAACGUACUUGCGAG ...(((((...(((((..((((((((((.......))))))))))..(((..(((((.(((.....)))))))).))))))))...)))))..(((((((((....)))))))))... ( -50.70, z-score = -2.06, R) >consensus CUGCUGUGGCAGCUCGAGCAAGUGUGUAAGUGCUGUGCGUAUGCGGACGACCUGCUCAUCUUGGUUGAGGGUCAGUCGCGAACUGAGAUAGAGGCAAAUGCCGCAAAGU_________ .(((..((((((.(((.((((((((((.......))))))))))...))).)))))).....(((((.....))))))))..........(((((....))))).............. (-14.24 = -17.30 + 3.06)

| Location | 24,157,030 – 24,157,132 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 61.76 |
| Shannon entropy | 0.63585 |
| G+C content | 0.50451 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -16.45 |
| Energy contribution | -18.95 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24157030 102 + 24543557 ACGCGCUGAAACCACACAGUGGCUCCAGACGCUGGAGAAGUAAGUCCAUCAUAA-GGUUCCAAAUAAUUGAACCGCCGAUGGAUGCCUGACCUCGUGUUACCA-------------- ((((((((........))))((((((((...))))).......(((((((....-(((((.........)))))...)))))))))).......)))).....-------------- ( -29.70, z-score = -1.78, R) >droAna3.scaffold_9904 1488 113 - 1642 ACGCACAACACUGCCACGCUUGCUCGAGCUGCCACAGCAGGGGAUCCAUCAUGAUGGUCCUUUUUAAUUGUACCAUCGAUGGAACACUGUGGAGUAUCCAUAAUCAUUAUCUU---- .........(((.((((.....(((..((((...)))).)))..((((((..((((((.(.........).)))))))))))).....)))))))..................---- ( -28.80, z-score = -0.70, R) >droVir3.scaffold_9533 3830 114 - 3988 ACGCACAACAUUUACACAUCUGCUCAAGCUGCACCAGCAAGGGGUCCAUCAUGA-GGUUCCAAAUGAAUGGGCCACAGAUAGAUCCCUGCGG-GCAGCCACAAAC-ACAUUUACGCU ..(((...............)))....(((((.((.(((.((((((.(((.((.-(((.(((......)))))).))))).)))))))))))-))))).......-........... ( -35.56, z-score = -2.63, R) >droGri2.scaffold_15056 4846 115 + 38174 ACGCACAGCACUUGCAUACACGCUCGAGCUGCCAUAGCAAGGUAUCCAUCAUGA-GGUUCCAUAUAAAUGGACCACAGAUGGAACCCUGCGG-GCAGCCACGAACCACAUUCAGGCU ..((...((....))........(((.((((((...(((.(((.((((((.((.-(((.((((....))))))).))))))))))).))).)-)))))..)))...........)). ( -44.20, z-score = -4.67, R) >consensus ACGCACAACACUUACACACUUGCUCGAGCUGCCACAGCAAGGGAUCCAUCAUGA_GGUUCCAAAUAAAUGGACCACAGAUGGAACCCUGCGG_GCAGCCACAAAC_ACAUUUA____ ...........................(((((.((....(((((((((((.....(((((.........)))))...)))))))))))..)).)))))................... (-16.45 = -18.95 + 2.50)


| Location | 24,157,030 – 24,157,132 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 61.76 |
| Shannon entropy | 0.63585 |
| G+C content | 0.50451 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -15.70 |
| Energy contribution | -18.33 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.997020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24157030 102 - 24543557 --------------UGGUAACACGAGGUCAGGCAUCCAUCGGCGGUUCAAUUAUUUGGAACC-UUAUGAUGGACUUACUUCUCCAGCGUCUGGAGCCACUGUGUGGUUUCAGCGCGU --------------(((......(((((.(((..(((((((..(((((.........)))))-...))))))))))))))).)))(((.((((((((((...))))))))))))).. ( -39.80, z-score = -3.28, R) >droAna3.scaffold_9904 1488 113 + 1642 ----AAGAUAAUGAUUAUGGAUACUCCACAGUGUUCCAUCGAUGGUACAAUUAAAAAGGACCAUCAUGAUGGAUCCCCUGCUGUGGCAGCUCGAGCAAGCGUGGCAGUGUUGUGCGU ----.........((((((((((((....)))).))))).))).(((((((......(((((((....)))).)))...(((((.((.(((......))))).)))))))))))).. ( -32.10, z-score = -0.49, R) >droVir3.scaffold_9533 3830 114 + 3988 AGCGUAAAUGU-GUUUGUGGCUGC-CCGCAGGGAUCUAUCUGUGGCCCAUUCAUUUGGAACC-UCAUGAUGGACCCCUUGCUGGUGCAGCUUGAGCAGAUGUGUAAAUGUUGUGCGU (((((..(((.-.(((((((((((-((((((((.((((((((.((.(((......)))..))-.)).)))))).))).))).)).))))))...)))))..)))..)))))...... ( -44.40, z-score = -3.04, R) >droGri2.scaffold_15056 4846 115 - 38174 AGCCUGAAUGUGGUUCGUGGCUGC-CCGCAGGGUUCCAUCUGUGGUCCAUUUAUAUGGAACC-UCAUGAUGGAUACCUUGCUAUGGCAGCUCGAGCGUGUAUGCAAGUGCUGUGCGU .(((.......)))(((.((((((-(.(((((((((((((((.(((((((....)))).)))-.)).)))))).)))))))...))))))))))(((..((.((....))))..))) ( -54.20, z-score = -4.75, R) >consensus ____UAAAUGU_GUUUGUGGCUGC_CCGCAGGGAUCCAUCGGUGGUCCAAUUAUAUGGAACC_UCAUGAUGGACCCCCUGCUGUGGCAGCUCGAGCAAGUGUGUAAGUGCUGUGCGU ...........................((((((((((((((..((((((......)))).))....))))))).)))))))....(((((.(..((......))..).))))).... (-15.70 = -18.33 + 2.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:57 2011