| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,099,761 – 24,099,911 |
| Length | 150 |
| Max. P | 0.997358 |

| Location | 24,099,761 – 24,099,911 |
|---|---|
| Length | 150 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Shannon entropy | 0.20288 |
| G+C content | 0.53210 |
| Mean single sequence MFE | -59.33 |
| Consensus MFE | -44.23 |
| Energy contribution | -43.63 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
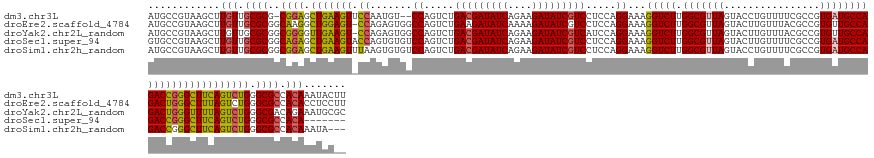
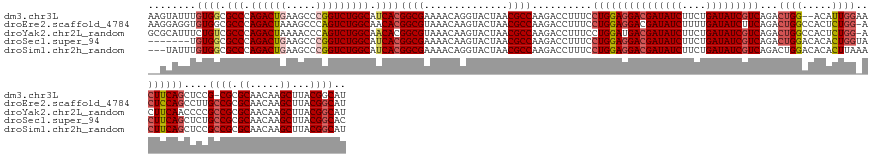
>dm3.chr3L 24099761 150 + 24543557 AUGCCGUAAGCUUGUUGCGCG-CGGAGCUGAAGUUCCAAUGU--CCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAAAUACUU .....(((...((((.((((.-((((.(((((((((.....(--((.....(((((((((....))))))))).....)))..(((((.(((((((................))))))))))))))))))))))))).)))).)))).))).. ( -60.89, z-score = -3.87, R) >droEre2.scaffold_4784 25390242 152 + 25762168 AUGCCGUAAGCUUGUUGCGCGGCAAGGCUGGAGU-CCAGAGUGGCCAGUCUGAAGAUAUCAAAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUACGCCGUGUUGCCAGACUGGGCUUUAGUCUGGGCGCCACACCUCCUU .((((((..((.....))))))))(((.(((.((-(((((.((.((((((((..(((((...(((((....(((....)))...)))))(((((............))))))))))..)))))))).)...).))))))).)))..))).... ( -61.50, z-score = -2.85, R) >droYak2.chr2L_random 2366577 152 + 4064425 AUGCCGUAAGCUUGUUGCGCGGCGGGGUUGAAGU-CCAGAGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCAUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUACGCCGUGUUGCCAGACUGGGUUUUAGUCUGGGCGACAGAAAUGCGC ..(((((..((.....)))))))(.(.(((..((-(((((.(((((((((((((((((((....)))))))))..........(((..((((((............))))))....))))))))))...))).)))))))..)))...).).. ( -59.70, z-score = -2.86, R) >droSec1.super_94 48640 146 + 93855 GUGCCGUAAGCUUGUUGCGCGGCAGAGCUGAAGUACCAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACA------- .((((((..((.....))))))))..((((......))))((((((((.(((((((((((....)))))))(((....)))..(((((.(((((((................)))))))))))).....)))).))))))))....------- ( -57.89, z-score = -2.58, R) >droSim1.chr2h_random 2494563 150 - 3178526 AUGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGUUUAAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAAAUA--- .((((((..((.....))))))))((((....))))..((((((((((.(((((((((((....)))))))(((....)))..(((((.(((((((................)))))))))))).....)))).))))))).))).....--- ( -56.69, z-score = -2.44, R) >consensus AUGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGU_CCAGUGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAAAUAC__ .((((....((.....))....((((.(((((((.((.......((.....(((((((((....))))))))).....))...(((((.(((((((................)))))))))))))))))))))))))))))............ (-44.23 = -43.63 + -0.60)
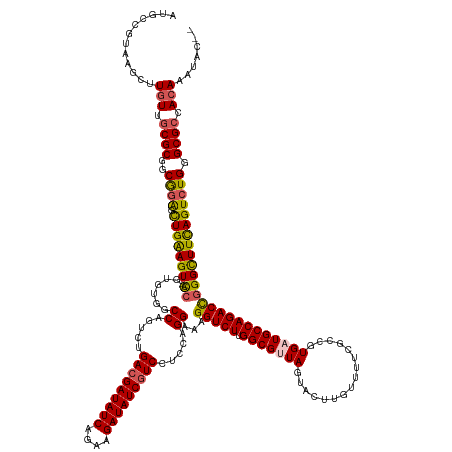
| Location | 24,099,761 – 24,099,911 |
|---|---|
| Length | 150 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Shannon entropy | 0.20288 |
| G+C content | 0.53210 |
| Mean single sequence MFE | -54.04 |
| Consensus MFE | -43.42 |
| Energy contribution | -44.62 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
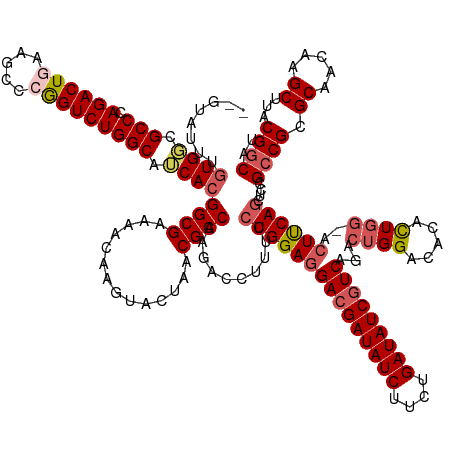
>dm3.chr3L 24099761 150 - 24543557 AAGUAUUUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGG--ACAUUGGAACUUCAGCUCCG-CGCGCAACAAGCUUACGGCAU .(((((((((..(((((((((((.....))))))).......))))...)))))))))...(((............(((((((((((((....)))))))))...((((--(.........))))))))).-.((.......))....))).. ( -53.61, z-score = -2.74, R) >droEre2.scaffold_4784 25390242 152 - 25762168 AAGGAGGUGUGGCGCCCAGACUAAAGCCCAGUCUGGCAACACGGCGUAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUUUGAUAUCUUCAGACUGGCCACUCUGG-ACUCCAGCCUUGCCGCGCAACAAGCUUACGGCAU (((((((((((..(((.(((((.......))))))))..)))(((((............)))))...))))...((((((...((((((....))))))((((((........)))))-)))))))))))((((.((.....))...)))).. ( -55.60, z-score = -3.00, R) >droYak2.chr2L_random 2366577 152 - 4064425 GCGCAUUUCUGUCGCCCAGACUAAAACCCAGUCUGGCAACACGGCGUAAACAAGUACUAACGCCAAGACCUUUCCUGGAUGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACUCUGG-ACUUCAACCCCGCCGCGCAACAAGCUUACGGCAU ((((.....(((.(((.(((((.......)))))))).))).((((.....((((.(((..((((...((......)).((((((((((....))))))))))...)))).....)))-))))......)))))))).....((.....)).. ( -51.60, z-score = -4.62, R) >droSec1.super_94 48640 146 - 93855 -------UGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGGUACUUCAGCUCUGCCGCGCAACAAGCUUACGGCAC -------(((.((((.((((((((((.((((((((((......((........))......))).)))))..(((.(...(((((((((....)))))))))...).)))......))..)))))).))))..)))).))).((.....)).. ( -54.90, z-score = -3.61, R) >droSim1.chr2h_random 2494563 150 + 3178526 ---UAUUUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUUAAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCAU ---...((((.((((((((((((.....))))))))......((((((...((((.((.......)))))))))..(((((((((((((....)))))))))...(((((...........)))))))))))))))).))))((.....)).. ( -54.50, z-score = -3.99, R) >consensus __GUAUUUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGG_ACUUCAGCUCCGCCGCGCAACAAGCUUACGGCAU ........((((.(((.((((((.....))))))))).))))((((..............))))..........(((((((((((((((....)))))))))...((((.....))))..))))))....((((.((.....))...)))).. (-43.42 = -44.62 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:53 2011