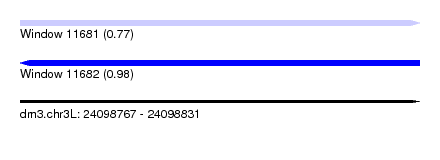
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,098,767 – 24,098,831 |
| Length | 64 |
| Max. P | 0.983623 |

| Location | 24,098,767 – 24,098,831 |
|---|---|
| Length | 64 |
| Sequences | 12 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 51.31 |
| Shannon entropy | 0.97553 |
| G+C content | 0.50526 |
| Mean single sequence MFE | -18.19 |
| Consensus MFE | -5.22 |
| Energy contribution | -4.74 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 2.29 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
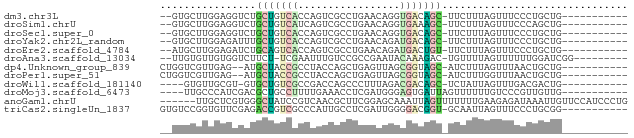
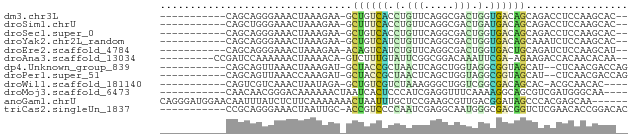
>dm3.chr3L 24098767 64 + 24543557 --GUGCUUGGAGGUCUGCUGUCACCAGUCGCCUGAACAGGUGACAGC-UUCUUUAGUUUCCCUGCUG----------- --..((..(((((.(((((((((((..((....))...)))))))))-......)))))))..))..----------- ( -22.60, z-score = -1.68, R) >droSim1.chrU 9462975 64 + 15797150 --GUGCUUGGAGGUCUGCUGUCAUCAGUCGCCUGAACAGGUGAAAGC-UUCUUUAGUUUCCCAGCUG----------- --..(((.(((((.((((((....)))((((((....))))))....-.....)))))))).)))..----------- ( -18.50, z-score = -0.73, R) >droSec1.super_0 20530130 64 - 21120651 --GUGCUUGGAGGUCUGCUGUCACCAGUCGCCUGAACAGGUGACAGC-UUCUUUAGUUUCCCUGCUG----------- --..((..(((((.(((((((((((..((....))...)))))))))-......)))))))..))..----------- ( -22.60, z-score = -1.68, R) >droYak2.chr2L_random 2365578 64 + 4064425 --GUGCUUGGAGAUUUGCUGUCACCAGUCGCCUGAACAGAUGACAGC-UUCUUUAGUUUCCCUGCUG----------- --..((..(((((((.(((((((.(..((....))...).)))))))-......)))))))..))..----------- ( -18.90, z-score = -1.81, R) >droEre2.scaffold_4784 25389499 64 + 25762168 --AUGCUUGGAGAUCUGCAGUCACCAGUCGCCUGAACAGAUGACUGU-UUCUUUAGUUUCCCUGCUG----------- --..((..(((((.(((.((....((((((.((....)).)))))).-..)).))))))))..))..----------- ( -18.40, z-score = -1.90, R) >droAna3.scaffold_13034 1066267 65 - 2223656 --UUGUGUUGUGGUCUUCU-UCGAAUUUGUCCGCCGAAUACAAAGAC-UGUUUUAGUUUUUUGGAUCGG--------- --(((((((((((.(....-........).))))..)))))))...(-((.(((((....))))).)))--------- ( -10.60, z-score = 0.21, R) >dp4.Unknown_group_839 5067 64 - 8057 CUGGUCGUUGAG--AUGCUACCGCCUACCAGCUGAGUUAGCGGUAGC-AUCUUUAGUUUAACUGCUG----------- ..(((.((((((--((((((((((..(((....).))..))))))))-)))))......))).))).----------- ( -25.50, z-score = -3.66, R) >droPer1.super_51 303050 64 - 524598 CUGGUCGUUGAG--AUGCUACCGCCUACCAGCUGAGUUAGCGGUAGC-AUCUUUGGUUUAACUGCUG----------- ..(((.((((((--((((((((((..(((....).))..))))))))-)))))......))).))).----------- ( -25.50, z-score = -3.38, R) >droWil1.scaffold_181140 1390206 61 - 1556085 ----GUGUUGCGU-GUGCUGUCGCCGACCAGCCCUUUAGACGACAGC-UCUAUUAGUUUGACGACUG----------- ----..((((((.-(.((((((((..(........)..).)))))))-.)........)).))))..----------- ( -12.40, z-score = 0.36, R) >droMoj3.scaffold_6473 13176143 63 + 16943266 ----UUGCCCAUCGACGCUGCCUUUUGAAACCUCGAUGGGAGUGAUUAGUUUUUUGUCCCGUUGUUG----------- ----...((((((((.(.............).))))))))...........................----------- ( -11.52, z-score = -0.32, R) >anoGam1.chrU 44970909 72 + 59568033 ------UUGCUCGUGGGCUAUCCGUCAACGCUUCGGAGCAAAUUAGUUUUUUUGAAGAGAUAAAUUGUUCCAUCCCUG ------.....(((.(((.....))).)))....(((((((.((.((((((....)))))).)))))))))....... ( -14.00, z-score = -0.16, R) >triCas2.singleUn_1837 3954 66 - 7082 GUGUCCGGUGUUCGAGACCGUCGCCCAUUGCCUCGAUUGGGGACGGU-GCAAUUAGUUUCCCUGCGG----------- ....((..........((((((.((((..........))))))))))-(((...........)))))----------- ( -17.80, z-score = 0.90, R) >consensus __GUGCGUGGAGGUCUGCUGUCACCAGUCGCCUGAACAGGUGACAGC_UUCUUUAGUUUCCCUGCUG___________ ................(((((((.................)))))))............................... ( -5.22 = -4.74 + -0.48)
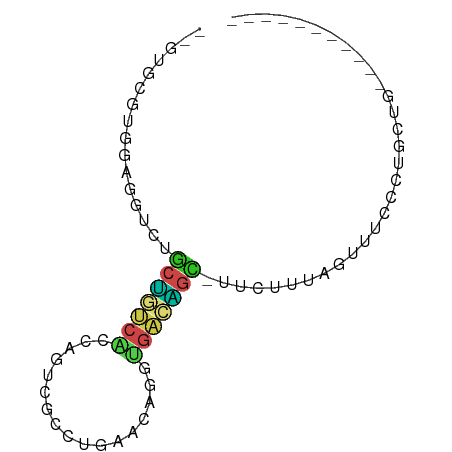
| Location | 24,098,767 – 24,098,831 |
|---|---|
| Length | 64 |
| Sequences | 12 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 51.31 |
| Shannon entropy | 0.97553 |
| G+C content | 0.50526 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -4.89 |
| Energy contribution | -4.05 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 2.60 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 24098767 64 - 24543557 -----------CAGCAGGGAAACUAAAGAA-GCUGUCACCUGUUCAGGCGACUGGUGACAGCAGACCUCCAAGCAC-- -----------..((.((....))......-(((((((((.(((.....))).)))))))))..........))..-- ( -24.30, z-score = -2.78, R) >droSim1.chrU 9462975 64 - 15797150 -----------CAGCUGGGAAACUAAAGAA-GCUUUCACCUGUUCAGGCGACUGAUGACAGCAGACCUCCAAGCAC-- -----------..(((((....))......-(((.(((.....((((....))))))).))).........)))..-- ( -15.70, z-score = -0.55, R) >droSec1.super_0 20530130 64 + 21120651 -----------CAGCAGGGAAACUAAAGAA-GCUGUCACCUGUUCAGGCGACUGGUGACAGCAGACCUCCAAGCAC-- -----------..((.((....))......-(((((((((.(((.....))).)))))))))..........))..-- ( -24.30, z-score = -2.78, R) >droYak2.chr2L_random 2365578 64 - 4064425 -----------CAGCAGGGAAACUAAAGAA-GCUGUCAUCUGUUCAGGCGACUGGUGACAGCAAAUCUCCAAGCAC-- -----------..((.((....))......-(((((((((.(((.....))).)))))))))..........))..-- ( -21.70, z-score = -2.82, R) >droEre2.scaffold_4784 25389499 64 - 25762168 -----------CAGCAGGGAAACUAAAGAA-ACAGUCAUCUGUUCAGGCGACUGGUGACUGCAGAUCUCCAAGCAU-- -----------..((.((....))......-.((((((((.(((.....))).))))))))...........))..-- ( -17.30, z-score = -1.24, R) >droAna3.scaffold_13034 1066267 65 + 2223656 ---------CCGAUCCAAAAAACUAAAACA-GUCUUUGUAUUCGGCGGACAAAUUCGA-AGAAGACCACAACACAA-- ---------((((....(((.(((.....)-)).)))....))))((((....)))).-.................-- ( -6.70, z-score = 0.24, R) >dp4.Unknown_group_839 5067 64 + 8057 -----------CAGCAGUUAAACUAAAGAU-GCUACCGCUAACUCAGCUGGUAGGCGGUAGCAU--CUCAACGACCAG -----------...............((((-(((((((((.(((.....))).)))))))))))--)).......... ( -24.80, z-score = -4.41, R) >droPer1.super_51 303050 64 + 524598 -----------CAGCAGUUAAACCAAAGAU-GCUACCGCUAACUCAGCUGGUAGGCGGUAGCAU--CUCAACGACCAG -----------...............((((-(((((((((.(((.....))).)))))))))))--)).......... ( -24.80, z-score = -4.53, R) >droWil1.scaffold_181140 1390206 61 + 1556085 -----------CAGUCGUCAAACUAAUAGA-GCUGUCGUCUAAAGGGCUGGUCGGCGACAGCAC-ACGCAACAC---- -----------...................-(((((((((.............)))))))))..-.........---- ( -15.12, z-score = -0.93, R) >droMoj3.scaffold_6473 13176143 63 - 16943266 -----------CAACAACGGGACAAAAAACUAAUCACUCCCAUCGAGGUUUCAAAAGGCAGCGUCGAUGGGCAA---- -----------...........................((((((((.(((((....)).))).))))))))...---- ( -16.50, z-score = -2.78, R) >anoGam1.chrU 44970909 72 - 59568033 CAGGGAUGGAACAAUUUAUCUCUUCAAAAAAACUAAUUUGCUCCGAAGCGUUGACGGAUAGCCCACGAGCAA------ .(((((((((....)))))))))..............((((((....((...........))....))))))------ ( -13.90, z-score = -0.46, R) >triCas2.singleUn_1837 3954 66 + 7082 -----------CCGCAGGGAAACUAAUUGC-ACCGUCCCCAAUCGAGGCAAUGGGCGACGGUCUCGAACACCGGACAC -----------(((((((....))...)))-((((((((((..........)))).))))))..........)).... ( -21.40, z-score = -1.24, R) >consensus ___________CAGCAGGGAAACUAAAGAA_GCUGUCACCUAUUCAGGCGACUGGCGACAGCAGACCUCCAAGCAC__ ................................((((((.(.(((.....))).).))))))................. ( -4.89 = -4.05 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:52 2011