| Sequence ID | dm3.chr3L |
|---|---|
| Location | 24,090,780 – 24,090,858 |
| Length | 78 |
| Max. P | 0.953954 |

| Location | 24,090,780 – 24,090,858 |
|---|---|
| Length | 78 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 65.72 |
| Shannon entropy | 0.67920 |
| G+C content | 0.50381 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -9.43 |
| Energy contribution | -10.56 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
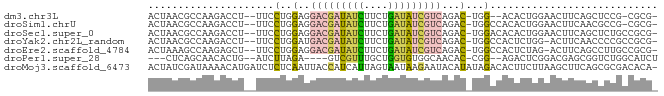
>dm3.chr3L 24090780 78 + 24543557 -CGCG-CGGAGCUGAAGUUCCAGUGU--CCA-GUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAA--AGGUCUUGGCGUUAGU -.(((-((((((....))))).))))--...-....(((((((((....)))))))))(.((((((.--...)))))).)..... ( -27.90, z-score = -2.70, R) >droSim1.chrU 10451082 80 - 15797150 -CGCG-CGGCGUUGAAGUUCCAGUGUGGCCA-GUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAA--AGGUCUUGGCGUUAGU -.(((-(.((((((......))))))((((.-.((((((((((((....))))))))).....))).--.))))...)))).... ( -26.20, z-score = -1.64, R) >droSec1.super_0 20526889 81 - 21120651 -CGCGGCAGAGCUGAAGUUCCAGUGUGUCCA-GUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAA--AGGUCUUGGCGUUAGU -(((((((..((((......)))).))))..-....(((((((((....)))))))))...(((((.--...))))))))..... ( -26.30, z-score = -2.06, R) >droYak2.chr2L_random 2363028 80 + 4064425 -CGCGGCGGGGUUGAAGU-CCAGAGUGGCCA-GUCUGACGAUAUCAGAAGAUAUCGUCAUCCAGGAA--AGGUCUUGGCGUUAGU -.(((.((((..(....(-(((((.(....)-.)))(((((((((....))))))))).....))).--.)..)))).))).... ( -27.70, z-score = -2.38, R) >droEre2.scaffold_4784 25147695 80 + 25762168 -CGCGGCAAGGCUGAAGU-CUAGAGUGGCCA-GUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAA--AGCUCUUGGCUUUAGU -.........((((((((-(.(((((..((.-....(((((((((....))))))))).....))..--.))))).))))))))) ( -33.00, z-score = -3.46, R) >droPer1.super_28 569058 73 - 1111753 AGAUGCCAGACCGCUCGUCCGAGUCU--CCG-GUGUUGCCACACCAGCAAACGAC----UCUAAGAU--CAGUGUUGCUGAG--- ......(((..((((.(((.(((((.--..(-((((....))))).(....))))----))...)))--.))))...)))..--- ( -19.50, z-score = -0.50, R) >droMoj3.scaffold_6473 13172871 84 + 16943266 -UGUGUCGCGCUGAAGCUUAAGAAGUGUCUAUAUGUAUUCUUAUUACUAAUGAUGGUAAUUGAGAGAUCAUGUUUUAUCGAUAGU -..(((((((((...........)))))..(((((..((((((((((((....)))))).))))))..)))))......)))).. ( -16.10, z-score = -0.18, R) >consensus _CGCGGCGGAGCUGAAGUUCCAGUGUGGCCA_GUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAA__AGGUCUUGGCGUUAGU ....................................(((((((((....)))))))))..((((((......))))))....... ( -9.43 = -10.56 + 1.13)
| Location | 24,090,780 – 24,090,858 |
|---|---|
| Length | 78 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 65.72 |
| Shannon entropy | 0.67920 |
| G+C content | 0.50381 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -7.55 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
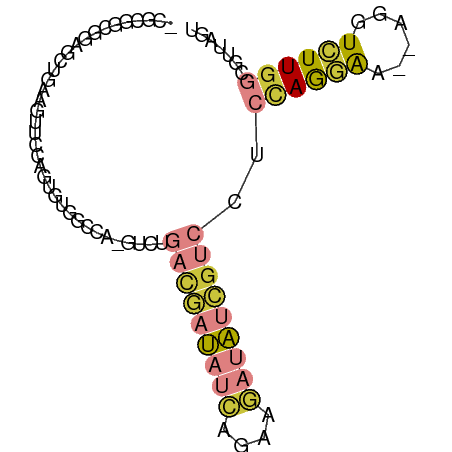
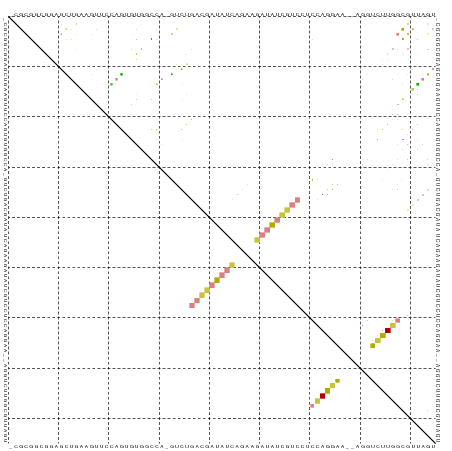
>dm3.chr3L 24090780 78 - 24543557 ACUAACGCCAAGACCU--UUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGAC-UGG--ACACUGGAACUUCAGCUCCG-CGCG- .....(((........--.(((.(...(((((((((....)))))))))...)-.))--)....(((.(....).))))-))..- ( -23.00, z-score = -2.33, R) >droSim1.chrU 10451082 80 + 15797150 ACUAACGCCAAGACCU--UUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGAC-UGGCCACACUGGAACUUCAACGCCG-CGCG- ......((((...((.--.....))..(((((((((....)))))))))....-))))................(((..-.)))- ( -21.20, z-score = -1.89, R) >droSec1.super_0 20526889 81 + 21120651 ACUAACGCCAAGACCU--UUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGAC-UGGACACACUGGAACUUCAGCUCUGCCGCG- .....(((..(((...--...(((((((((((((((....)))))))))...(-..(.....)..)..)))))).)))...)))- ( -24.50, z-score = -2.94, R) >droYak2.chr2L_random 2363028 80 - 4064425 ACUAACGCCAAGACCU--UUCCUGGAUGACGAUAUCUUCUGAUAUCGUCAGAC-UGGCCACUCUGG-ACUUCAACCCCGCCGCG- .....(((..(((...--..((.(..((((((((((....))))))))))..)-.))....)))((-.........))...)))- ( -22.10, z-score = -3.33, R) >droEre2.scaffold_4784 25147695 80 - 25762168 ACUAAAGCCAAGAGCU--UUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGAC-UGGCCACUCUAG-ACUUCAGCCUUGCCGCG- ......((((((.(((--...(((((((((((((((....))))))))).(..-....).))))))-.....)))))))..)).- ( -26.90, z-score = -2.81, R) >droPer1.super_28 569058 73 + 1111753 ---CUCAGCAACACUG--AUCUUAGA----GUCGUUUGCUGGUGUGGCAACAC-CGG--AGACUCGGACGAGCGGUCUGGCAUCU ---....((...((((--.((((.((----(((.....(((((((....))))-)))--.))))).)).)).))))...)).... ( -25.60, z-score = -1.19, R) >droMoj3.scaffold_6473 13172871 84 - 16943266 ACUAUCGAUAAAACAUGAUCUCUCAAUUACCAUCAUUAGUAAUAAGAAUACAUAUAGACACUUCUUAAGCUUCAGCGCGACACA- ((((..(((......(((....)))......)))..))))..((((((.............)))))).((....))........- ( -6.22, z-score = 0.32, R) >consensus ACUAACGCCAAGACCU__UUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGAC_UGGCCACACUGGAACUUCAGCCCCGCCGCG_ ...........................(((((((((....))))))))).................................... ( -7.55 = -8.14 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:50 2011