| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,197,002 – 6,197,113 |
| Length | 111 |
| Max. P | 0.865768 |

| Location | 6,197,002 – 6,197,113 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.78 |
| Shannon entropy | 0.58357 |
| G+C content | 0.47150 |
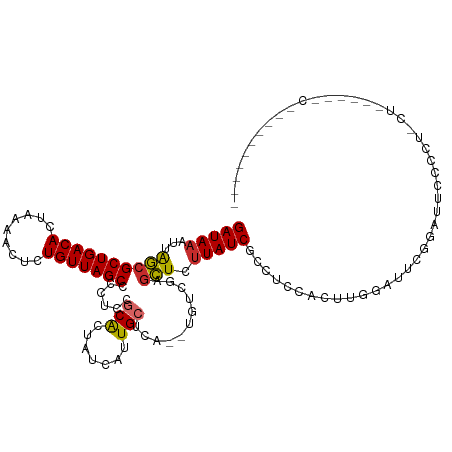
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -12.04 |
| Energy contribution | -11.67 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.08 |
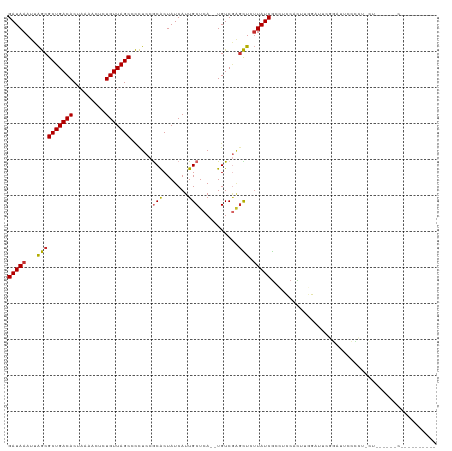
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.865768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
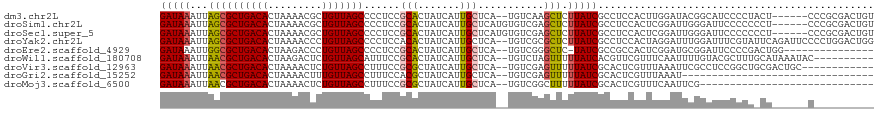
>dm3.chr2L 6197002 111 - 23011544 GAUAAAUUAGCGCUGACACUAAAACGCUGUUAGCCCCUCCGCACUAUCAUUGCUCA--UGUCAAGCUCUUAUCGCCUCCACUUGGAUACGGCAUCCCCUACU------CCCGCGACUGU (((((...(((((((((((......).)))))))......(((.......)))...--......))).)))))((((((....)))...)))..........------........... ( -21.90, z-score = -1.13, R) >droSim1.chr2L 5991533 113 - 22036055 GAUAAAUUAGCGCUGACACUAAAACGCUGUUAGCCCCUCCGCACUAUCAUUGCUCAUGUGUCGAGCUCUUAUCGCCUCCACUCGGAUUGGGAUUCCCCCCCU------CCCGCGACUGU (((((...(((((((((((......).)))))))...((.((((.............)))).))))).)))))((.(((....)))..((((.........)------)))))...... ( -25.72, z-score = -1.19, R) >droSec1.super_5 4268379 113 - 5866729 GAUAAAUUAGCGCUGACACUAAAACGCUGUUAGCCCCUCCGCACUAUCAUUGCUCAUGUGUCGAGCUCUUAUCGCCUCCACUCGGAUUGGGAUUCCCCCCCU------CCCGCGACUGU (((((...(((((((((((......).)))))))...((.((((.............)))).))))).)))))((.(((....)))..((((.........)------)))))...... ( -25.72, z-score = -1.19, R) >droYak2.chr2L 15612112 117 + 22324452 GAUAAAUUAGCGCUGACACUAAAACCCUGUUAGCCCCUCCACACUAUCAUUGCUCA--UGUCGCGCUCUUAUCGCCUCCACUAGGAUUUGGAUUUCGUAUUCAGAUUCCCCUGGACUGG (((((...((((((((((.........))))))).................((...--....))))).))))).((((((...((((((((((.....))))))))))...))))..)) ( -26.10, z-score = -1.69, R) >droEre2.scaffold_4929 15124337 101 - 26641161 GAUAAAUUGGCGCUGACACUAAGACCCUGUUAGCCCCUCCGCACUAUCAUUGCUCA--UGUCGGGCUC-UAUCGCCGCCACUCGGAUGCGGAUUCCCCGACUGG--------------- ........((.(((((((.........))))))).))...(((.......))).((--.((((((...-..((((..((....))..))))....)))))))).--------------- ( -28.50, z-score = -0.96, R) >droWil1.scaffold_180708 5041719 107 + 12563649 GAUAAAUUAACGCUGACACUAAGACUCUGUUAGCAUUUCCGCACUAUCAUUGCUCA--UGUCUAGUUUUUAUCACGUUCGUUUCAAUUUUGUACGCUUUGCAUAAAUAC---------- ((((((..((((((((((.........)))))))......(((.......)))...--......)))))))))............((((((((.....)))).))))..---------- ( -14.80, z-score = 0.22, R) >droVir3.scaffold_12963 17168745 105 + 20206255 GAUAAAUUAACGCUGACACUAAAACUCUGUUAGCCUUUCCGCGCUAUCAUUGCUCA--UGUCGAGUUUUUAUCGCACUCGUUUAAAUUCGCCUCCGGCUGCGACUGC------------ ((((((.....(((((((.........))))))).................((((.--....)))).))))))(((.((((........(((...))).)))).)))------------ ( -20.70, z-score = -0.52, R) >droGri2.scaffold_15252 14550706 85 + 17193109 GAUAAAUUAACGCUGACACUAAAACUUUGUUAGCCUUUCCACGCUAUCAUUGCUCA--UGUCGAGUUUUUAUCGCACUCGUUUAAAU-------------------------------- ((((((.....(((((((.........))))))).................((((.--....)))).))))))..............-------------------------------- ( -13.70, z-score = -1.26, R) >droMoj3.scaffold_6500 19001761 88 + 32352404 GAUAAAUUAACGCUGACACUAAAACUCUGUUAGCCUUUCCGCGCUAUCAUUGCUCA--UGUCGGCUUUUUAUCGCACUCGUUUCAAUUCG----------------------------- ((((((.....(((((((......(..((.((((........)))).))..)....--)))))))..)))))).................----------------------------- ( -16.30, z-score = -2.00, R) >consensus GAUAAAUUAGCGCUGACACUAAAACUCUGUUAGCCCCUCCGCACUAUCAUUGCUCA__UGUCGAGCUCUUAUCGCCUCCACUUGGAUUCGGAUUCCCCU_CU______C__________ (((((...((((((((((.........)))))))......(((.......)))...........))).))))).............................................. (-12.04 = -11.67 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:40 2011