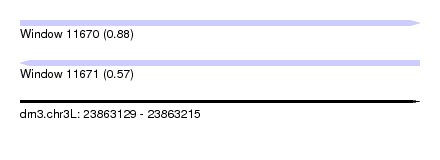
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,863,129 – 23,863,215 |
| Length | 86 |
| Max. P | 0.881777 |

| Location | 23,863,129 – 23,863,215 |
|---|---|
| Length | 86 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 58.13 |
| Shannon entropy | 0.73171 |
| G+C content | 0.56318 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -11.48 |
| Energy contribution | -13.28 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
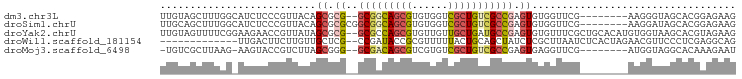
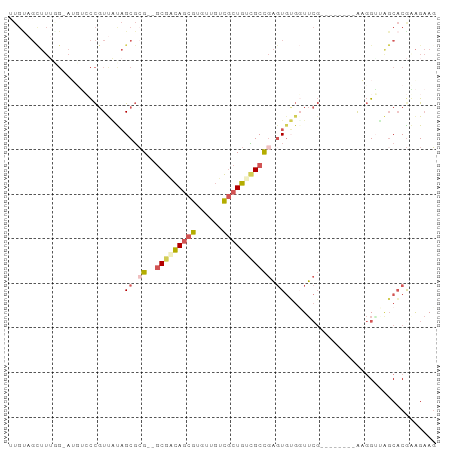
>dm3.chr3L 23863129 86 + 24543557 UUGUAGCUUUGGCAUCUCCCGUUACAGCGCG--GCGGCAGCGUGUGGUCGCUGUCGCCGAGUGUGGUUCG--------AAGGGUAGCACGGAGAAG .....((....)).((((((((....)))((--(((((((((......))))))))))).((((...((.--------...))..))))))))).. ( -38.70, z-score = -2.54, R) >droSim1.chrU 8524665 88 - 15797150 UUGCAGCUUUGGCAUCUCCCGUUACAGCGCGCGGCGGCAGCGUGUGGUCGCUGUCGCCGAGUGUGGUUCG--------AAGGAUAGCACGGAGAAG ......(((((((((((..((..((.((((.(((((((((((......))))))))))).)))).)).))--------..)))).)).)))))... ( -41.60, z-score = -2.59, R) >droYak2.chrU 7465418 94 + 28119190 UUGUAGUUUUCGGAAGAACCGUUAUAGCGCG--GCGCCAGCGUGUUGUUGCUGAUGCCGAGUGUGUUUCGCUGCACAUGUGGUAAGCACGUAGAAG .(((((..(((....)))...)))))((.((--(((.(((((......))))).))))).))((((((.((..(....)..))))))))....... ( -31.50, z-score = -0.84, R) >droWil1.scaffold_181154 230293 81 + 4610121 -------------UUGACUUCUUGUUGCUCG--CCGAUACCGCGUUUUUACUGCAGCUAUCUCGCUUAAUCUCACUAGAACGUUCCCUCGAGGCAG -------------............(((.(.--.(((....(((((((......(((......)))..........)))))))....))).)))). ( -11.69, z-score = 1.36, R) >droMoj3.scaffold_6498 344502 84 - 3408170 -UGUCGCUUAAG-AAGUACCGUCUUAGCGGG--GCGACAGCGUCGUGUCGCUGUCGCCGAGUGAGGUUCG--------AUGGUAGGCACAAAGAAU -..((((((...-.....((((....))))(--(((((((((......)))))))))))))))).((((.--------.((.......))..)))) ( -34.40, z-score = -2.84, R) >consensus UUGUAGCUUUGG_AUGUCCCGUUAUAGCGCG__GCGACAGCGUGUUGUCGCUGUCGCCGAGUGUGGUUCG________AAGGUUAGCACGAAGAAG ..........................((((...(((((((((......)))))))))...))))................................ (-11.48 = -13.28 + 1.80)

| Location | 23,863,129 – 23,863,215 |
|---|---|
| Length | 86 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 58.13 |
| Shannon entropy | 0.73171 |
| G+C content | 0.56318 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -8.14 |
| Energy contribution | -10.46 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
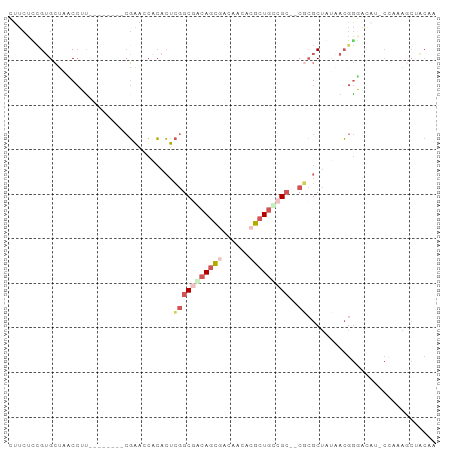
>dm3.chr3L 23863129 86 - 24543557 CUUCUCCGUGCUACCCUU--------CGAACCACACUCGGCGACAGCGACCACACGCUGCCGC--CGCGCUGUAACGGGAGAUGCCAAAGCUACAA ..(((((((.........--------...)).((((.(((((.(((((......))))).)))--)).).)))....))))).............. ( -25.60, z-score = -1.33, R) >droSim1.chrU 8524665 88 + 15797150 CUUCUCCGUGCUAUCCUU--------CGAACCACACUCGGCGACAGCGACCACACGCUGCCGCCGCGCGCUGUAACGGGAGAUGCCAAAGCUGCAA ..(((((((((.......--------.((.......))((((.(((((......))))).))))))))(......).)))))(((.......))). ( -27.90, z-score = -0.78, R) >droYak2.chrU 7465418 94 - 28119190 CUUCUACGUGCUUACCACAUGUGCAGCGAAACACACUCGGCAUCAGCAACAACACGCUGGCGC--CGCGCUAUAACGGUUCUUCCGAAAACUACAA .......((((........((((........))))...(((..((((........))))..))--))))).....(((.....))).......... ( -23.90, z-score = -1.44, R) >droWil1.scaffold_181154 230293 81 - 4610121 CUGCCUCGAGGGAACGUUCUAGUGAGAUUAAGCGAGAUAGCUGCAGUAAAAACGCGGUAUCGG--CGAGCAACAAGAAGUCAA------------- .(((((((..(((....)))..)))).....((..((((.((((.((....)))))))))).)--)..)))............------------- ( -18.60, z-score = 0.33, R) >droMoj3.scaffold_6498 344502 84 + 3408170 AUUCUUUGUGCCUACCAU--------CGAACCUCACUCGGCGACAGCGACACGACGCUGUCGC--CCCGCUAAGACGGUACUU-CUUAAGCGACA- ..................--------............((((((((((......)))))))))--).((((((((.......)-))).))))...- ( -27.00, z-score = -2.98, R) >consensus CUUCUCCGUGCUAACCUU________CGAACCACACUCGGCGACAGCGACAACACGCUGCCGC__CGCGCUAUAACGGGACAU_CCAAAGCUACAA .......((((...............(((.......)))(((((((((......)))))))))...)))).......................... ( -8.14 = -10.46 + 2.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:42 2011