| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,689,431 – 23,689,492 |
| Length | 61 |
| Max. P | 0.998822 |

| Location | 23,689,431 – 23,689,492 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 57.49 |
| Shannon entropy | 0.75631 |
| G+C content | 0.39342 |
| Mean single sequence MFE | -12.00 |
| Consensus MFE | -7.54 |
| Energy contribution | -7.18 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
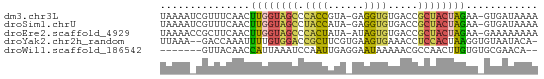
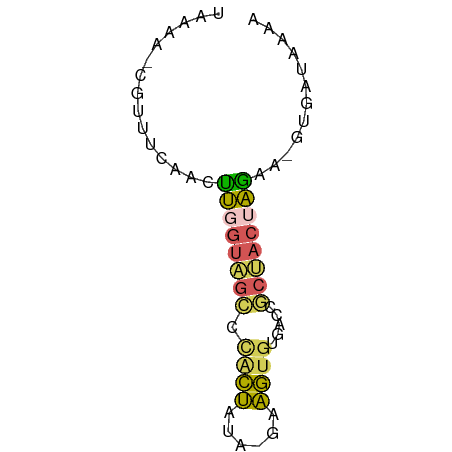
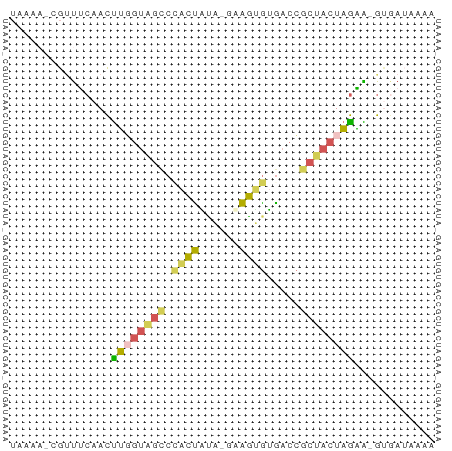
>dm3.chr3L 23689431 61 + 24543557 UAAAAUCGUUUCAACUUGGUAGCCCACCGUA-GAGGUGUGACCGCUACUAGAA-GUGAUAAAA ....((((((((....((((((((((((...-..)))).)...))))))))))-))))).... ( -16.50, z-score = -2.63, R) >droSim1.chrU 11407521 61 - 15797150 UAAAAUCGUUUCAACUUGGUAGCCUACCAUA-GAGGUGUGACCGCUACUAGAA-GUGAUAAAA ....((((((((....(((((((.((((...-..)))).....))))))))))-))))).... ( -13.70, z-score = -1.68, R) >droEre2.scaffold_4929 23377480 61 + 26641161 UAAAACCGCUUCAACUUGGUAGCCCACUAUA-AUAGUGUGACCGCUACUAGAA-GAAAAAAAA ........((((....(((((((((((((..-.))))).)...))))))))))-)........ ( -12.50, z-score = -2.82, R) >droYak2.chr2h_random 944802 60 - 3774259 UUAAA--GACCAAAUUUUGUGGACCGCUUCGUGAAGUGAAACCUCCACUAAGGUGUAAUACA- ....(--.(((.......(((((.(((((....))))).....)))))...))).)......- ( -10.40, z-score = -0.42, R) >droWil1.scaffold_186542 818 54 - 1333 -------GUUACAACCAUUAAAUCCAAUUGAGGAAUAAAAACGCCAACUUGUGUGCGAACA-- -------(((............(((......))).......(((((.....)).)))))).-- ( -6.90, z-score = -0.38, R) >consensus UAAAA_CGUUUCAACUUGGUAGCCCACUAUA_GAAGUGUGACCGCUACUAGAA_GUGAUAAAA ...............((((((((.((((......)))).....))))))))............ ( -7.54 = -7.18 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:37 2011