
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,187,882 – 6,187,982 |
| Length | 100 |
| Max. P | 0.537218 |

| Location | 6,187,882 – 6,187,982 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.85 |
| Shannon entropy | 0.55577 |
| G+C content | 0.49004 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.02 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6187882 100 + 23011544 --ACCCCAUUUUAAUGCUGUGCUGUCUCUGGCUUGAUUAGCGUCGCUGAAUUGACAGAAAUUGUCACCAGCCAGCCAGAGUGCGCUUCAACGGCAAACUCCC --............((((((((.((((((((((.(((....)))((((...((((((...)))))).)))).)))))))).))))....))))))....... ( -31.20, z-score = -2.33, R) >droSim1.chr2L 5980648 100 + 22036055 --ACCCCAUUUUAAUGCUGUGCUGUCUCUGGCUUGAUUAGCGUCGCUGAAUUGACAGAAAUUGUCACCAGCCAGCCAGAGUGUGCUUCAACGGCAAACUCCC --............((((((((...((((((((.(((....)))((((...((((((...)))))).)))).))))))))...))....))))))....... ( -31.80, z-score = -2.64, R) >droSec1.super_5 4259054 100 + 5866729 --ACCCCAUUUUAAUGCUGUGCUGUCUCUGGCUUGAUUAGCGUCGCUGAAUUGACAGAAAUUGUCACCAGCCAGCCAGAGUGUGCUUCAACGGCAAACUCCC --............((((((((...((((((((.(((....)))((((...((((((...)))))).)))).))))))))...))....))))))....... ( -31.80, z-score = -2.64, R) >droYak2.chr2L 15602621 96 - 22324452 --ACCCCAUUUUAAUGCUGUGCUGUCUCUGGCUUGAUUAGCGUCGCUGAAUUGACAGAAAUUGUCACCAGCCA----GAGUGUGCUUCAACAGCAAACUUCC --............((((((((...((((((((.(.(((((...)))))..((((((...))))))).)))))----)))...))....))))))....... ( -30.70, z-score = -3.09, R) >droEre2.scaffold_4929 15115450 95 + 26641161 --ACCCCAUUUUAAUGCUGUGCUGUCUCUGGCUUGAUUAGCGUCGCUGAAUUGACAGAAAUUGUCACCAGCCAGC-----CAACUCCCAACGCCCAACGCCC --.............((((.((((............(((((...)))))..((((((...)))))).))))))))-----...................... ( -17.90, z-score = -0.08, R) >droWil1.scaffold_180708 5028102 92 - 12563649 GGAGCCUGUUUAAAUGCUCGACUGUCUCUUGCUUGAUUAGCGCCGCUGAAUUGACAGAAAUUGUCACUAGCAGAGCCACAUGACAGCUGAUG---------- ..(((.((((.....((((...........(((.....)))...((((...((((((...)))))).)))).)))).....)))))))....---------- ( -22.70, z-score = -0.10, R) >droVir3.scaffold_12963 17155257 94 - 20206255 --ACCCUAUUUAAAUGCUCAACAGUCGCUCGCUUGAUUAGCGCCGCUGAAUUGACAGAAAUUGUCACUAGCAGGUCUAGAGUCACAGCAAGACCCC------ --............((((.....((.(((((((.....)))(((((((...((((((...)))))).)))).)))...)))).)))))).......------ ( -21.10, z-score = -0.69, R) >droMoj3.scaffold_6500 18984312 84 - 32352404 --ACCCUAUUUAAAAGCUGCACAGUCGCUCGCUUGAUUAGCGCCGCUGAAUUGACAGAAAUUGUCACAAGCAG-UUGGCACGCCCAC--------------- --............((((((.(((.((..((((.....)))).)))))...((((((...))))))...))))-))((....))...--------------- ( -21.50, z-score = -0.87, R) >droGri2.scaffold_15252 14537651 87 - 17193109 --UCCCCAUUUAAAUGCUCAUGAGUCGCUCGCUUGAUUAGCGCCGCUGAAUUGACAGAAAUUGUCACUAGCAGGUCUACAGUUCUAAUC------------- --.............(((.((((((.....)))).)).)))(((((((...((((((...)))))).)))).)))..............------------- ( -14.10, z-score = 1.09, R) >consensus __ACCCCAUUUUAAUGCUGUGCUGUCUCUGGCUUGAUUAGCGUCGCUGAAUUGACAGAAAUUGUCACCAGCCAGCCAGAAUGUCCUUCAACGGCAAAC__CC ............................(((((.(.(((((...)))))..((((((...))))))).)))))............................. (-11.06 = -11.02 + -0.04)

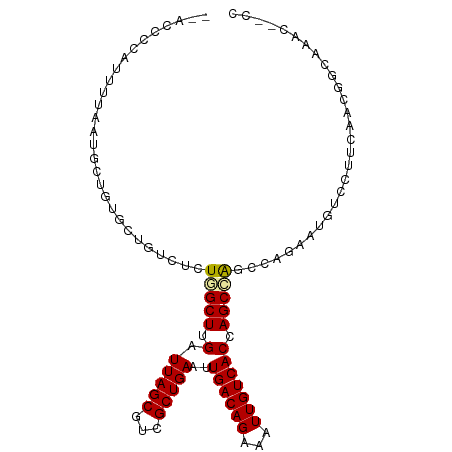
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:39 2011