| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,590,095 – 23,590,240 |
| Length | 145 |
| Max. P | 0.998240 |

| Location | 23,590,095 – 23,590,240 |
|---|---|
| Length | 145 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 50.00 |
| Shannon entropy | 0.70520 |
| G+C content | 0.47344 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.13 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
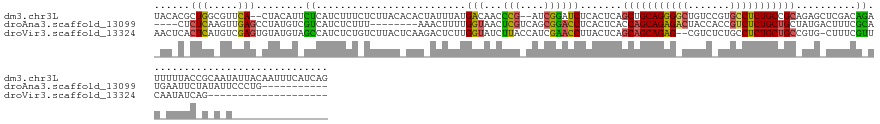
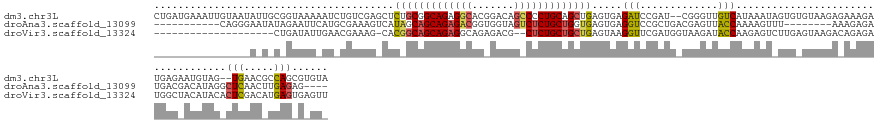
>dm3.chr3L 23590095 145 + 24543557 UACACGCUGGCGUUCA--CUACAUUCUCAUCUUUCUCUUACACACUAUUUAUGACAACCCG--AUCGGAUCUCACUCAGCUGCAGGGGCUGUCCGUGCCUCUGCCGCAGAGCUCGACAGAUUUUUACCGCAAUAUUACAAUUUCAUCAG .....((.((.((...--..))......((((.((.......................((.--...))......(((.((.((((((((.......)))))))).)).)))...)).)))).....))))................... ( -28.50, z-score = -0.47, R) >droAna3.scaffold_13099 1780302 126 - 3324263 ----CUCUCAAGUUGAGCCUAUGUCGUCAUCUCUUU--------AAACUUUUGGUAACUCGUCAGCGGACCUCACUCACCAGCAGAGACUACCACCGUCUCUGCUGCUAUGACUUUCGCAUGAAUUCUAUAUUCCCUG----------- ----..(((.....))).....((..(((.......--------.......)))..))...((((((((..(((.....((((((((((.......))))))))))...)))..))))).)))...............----------- ( -29.14, z-score = -2.19, R) >droVir3.scaffold_13324 1529453 126 - 2960039 AACUCACUCAUGUCGAGUGUAUGUAGCCAUCUCUGUCUUACUCAAGACUCUUGGUAUCUUACCAUCGAACCUUACUCAGCAGCAGAG--CGUCUCUGCCUCUGCUGCCGUG-CUUUCGUUCAAUAUCAG-------------------- .((.(((((.....)))))...)).((((.....(((((....)))))...))))...........((((......(.(((((((((--.((....))))))))))).)..-.....))))........-------------------- ( -34.82, z-score = -3.05, R) >consensus _AC_CACUCAAGUUGAG_CUAUGUAGUCAUCUCUCUCUUAC_CAAAACUUUUGGUAACUCG_CAUCGGACCUCACUCAGCAGCAGAG_CUGUCACUGCCUCUGCUGCAGUG_CUUUCGCAUAAUUUCAG_A_U________________ ......(((.....)))........((.........................(((...(((....)))))).......(((((((((((.......)))))))))))..........)).............................. (-16.79 = -16.13 + -0.65)
| Location | 23,590,095 – 23,590,240 |
|---|---|
| Length | 145 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 50.00 |
| Shannon entropy | 0.70520 |
| G+C content | 0.47344 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
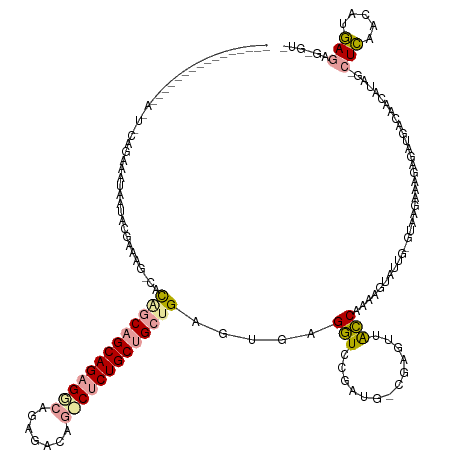
>dm3.chr3L 23590095 145 - 24543557 CUGAUGAAAUUGUAAUAUUGCGGUAAAAAUCUGUCGAGCUCUGCGGCAGAGGCACGGACAGCCCCUGCAGCUGAGUGAGAUCCGAU--CGGGUUGUCAUAAAUAGUGUGUAAGAGAAAGAUGAGAAUGUAG--UGAACGCCAGCGUGUA ...............(((((((......((((.((..((((.((.((((.(((.......))).)))).)).))))(((((((...--.))))).)).................)).)))).....)))))--)).(((....)))... ( -36.20, z-score = 0.35, R) >droAna3.scaffold_13099 1780302 126 + 3324263 -----------CAGGGAAUAUAGAAUUCAUGCGAAAGUCAUAGCAGCAGAGACGGUGGUAGUCUCUGCUGGUGAGUGAGGUCCGCUGACGAGUUACCAAAAGUUU--------AAAGAGAUGACGACAUAGGCUCAACUUGAGAG---- -----------((((((.......((((((((....)).....((((((((((.......))))))))))))))))....))).))).((((((.((....(((.--------........)))......))...))))))....---- ( -36.60, z-score = -1.85, R) >droVir3.scaffold_13324 1529453 126 + 2960039 --------------------CUGAUAUUGAACGAAAG-CACGGCAGCAGAGGCAGAGACG--CUCUGCUGCUGAGUAAGGUUCGAUGGUAAGAUACCAAGAGUCUUGAGUAAGACAGAGAUGGCUACAUACACUCGACAUGAGUGAGUU --------------------(((.((((((((....(-(.(((((((((((.(......)--))))))))))).))...))))))))......(((((((...)))).)))...)))..(((....))).(((((.....))))).... ( -40.30, z-score = -2.66, R) >consensus ________________A_U_CAGAAAUAAUACGAAAG_CACAGCAGCAGAGGCAGAGACAG_CUCUGCUGCUGAGUGAGGUCCGAUG_CGAGUUACCAAAAGUAUUG_GUAAGAAAGAGAUGACAACAUAG_CUCAACAUGAGAG_GU_ ........................................(((((((((((((.......))))))))))))).....((((((....)))...)))...................................(((.....)))...... (-17.30 = -18.20 + 0.90)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:31 2011