
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,565,072 – 23,565,179 |
| Length | 107 |
| Max. P | 0.999875 |

| Location | 23,565,072 – 23,565,179 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.64 |
| Shannon entropy | 0.45477 |
| G+C content | 0.34161 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -23.90 |
| Energy contribution | -25.58 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.27 |
| Mean z-score | -5.47 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.62 |
| SVM RNA-class probability | 0.999863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
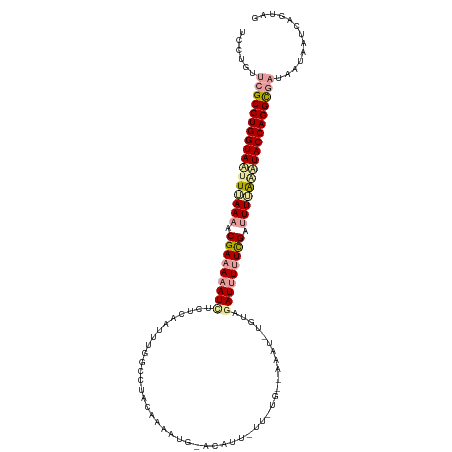
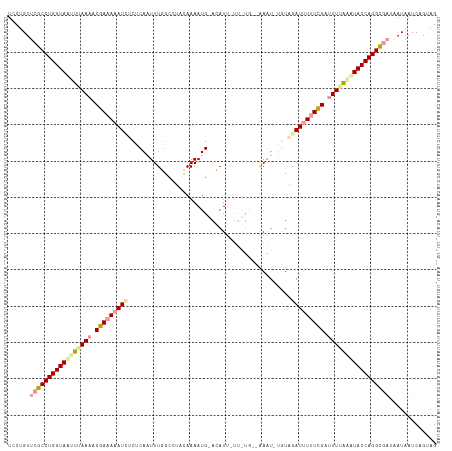
>dm3.chr3L 23565072 107 + 24543557 UUCUGUUCGCCUGGUACUUGAAAUCGAAAAAUCACUCA-UUUGACCGCUAAAAUG-ACAU---------AACUUAGUCAAUUUAUUGUUUUUGUGUACCAGGUUUAAAUAAUCUGUAG ........(((((((((.(((((.(((....(((....-..)))....((((.((-((..---------......)))).))))))).))))).)))))))))............... ( -22.10, z-score = -2.32, R) >droEre2.scaffold_4820 291585 117 + 10470090 UCCUGUUCGCCUGGUAAUUUAAAACGAAAAAUUUCACAAUUUUGCCUACAAAAUGUAAAUUAUUAUGGCGAAU-UGUAGAUUUUUCGGAUUGAAUUACCAGGCGAUCAUAAUCAGCAG ...((.((((((((((((((((..((((((((((.(((((((.(((.....(((....))).....)))))))-)))))))))))))..)))))))))))))))).)).......... ( -46.80, z-score = -8.86, R) >droSec1.super_141 1621 118 + 69287 UCCAGUUCGCCUGGUAGUUUAAACCGAAAAAUCUCUCAAUUUGGCAUACAAAAUGCACAUUUUUGUGCAAAAUCUUUGGAUCUUUCGAUUUUAGAUACCAGGCGAUGAUAAUCAGUAG ......((((((((((.((((((.(((((.((((.....((((.....)))).((((((....))))))........)))).)))))..))))))))))))))))............. ( -37.10, z-score = -5.22, R) >consensus UCCUGUUCGCCUGGUAAUUUAAAACGAAAAAUCUCUCAAUUUGGCCUACAAAAUG_ACAUU_UU_UG__AAAU_UGUAGAUUUUUCGAUUUUAAAUACCAGGCGAUAAUAAUCAGUAG ......(((((((((((((((((.(((((((((.............................................))))))))).)))))))))))))))))............. (-23.90 = -25.58 + 1.68)

| Location | 23,565,072 – 23,565,179 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.64 |
| Shannon entropy | 0.45477 |
| G+C content | 0.34161 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -17.10 |
| Energy contribution | -19.21 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.83 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
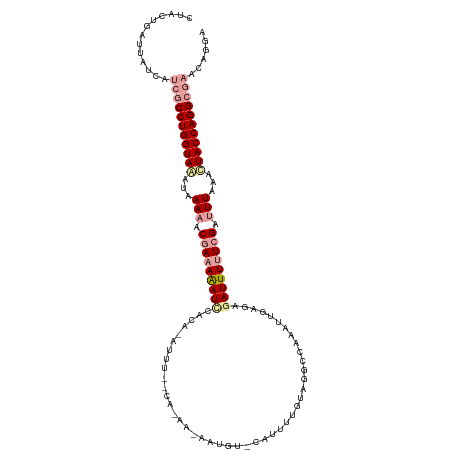
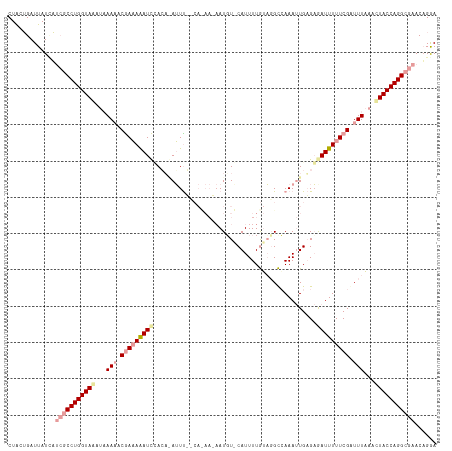
>dm3.chr3L 23565072 107 - 24543557 CUACAGAUUAUUUAAACCUGGUACACAAAAACAAUAAAUUGACUAAGUU---------AUGU-CAUUUUAGCGGUCAAA-UGAGUGAUUUUUCGAUUUCAAGUACCAGGCGAACAGAA ................((((((((..............((((((..(((---------(...-.....)))))))))).-(((.(((....)))...))).))))))))......... ( -23.00, z-score = -2.22, R) >droEre2.scaffold_4820 291585 117 - 10470090 CUGCUGAUUAUGAUCGCCUGGUAAUUCAAUCCGAAAAAUCUACA-AUUCGCCAUAAUAAUUUACAUUUUGUAGGCAAAAUUGUGAAAUUUUUCGUUUUAAAUUACCAGGCGAACAGGA ..........((.(((((((((((((.((..((((((((.((((-(((.(((((((...........)))).)))..)))))))..))))))))..)).))))))))))))).))... ( -40.00, z-score = -6.61, R) >droSec1.super_141 1621 118 - 69287 CUACUGAUUAUCAUCGCCUGGUAUCUAAAAUCGAAAGAUCCAAAGAUUUUGCACAAAAAUGUGCAUUUUGUAUGCCAAAUUGAGAGAUUUUUCGGUUUAAACUACCAGGCGAACUGGA .............((((((((((....(((((((((((((....(((((.(((..(((((....)))))...))).)))))....)))))))))))))....))))))))))...... ( -39.60, z-score = -5.65, R) >consensus CUACUGAUUAUCAUCGCCUGGUAAAUAAAAACGAAAAAUCCACA_AUUU__CA_AA_AAUGU_CAUUUUGUAGGCCAAAUUGAGAGAUUUUUCGAUUUAAACUACCAGGCGAACAGGA .............(((((((((((...(((.(((((((((.............................................))))))))).)))...)))))))))))...... (-17.10 = -19.21 + 2.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:30 2011