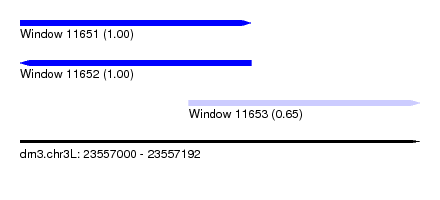
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,557,000 – 23,557,192 |
| Length | 192 |
| Max. P | 0.999334 |

| Location | 23,557,000 – 23,557,111 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.59 |
| Shannon entropy | 0.63985 |
| G+C content | 0.45874 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -19.03 |
| Energy contribution | -18.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 23557000 111 + 24543557 ----AAAGUGCUGGCGCGGCAGUUCU-GGUUUCUUUUGCGGGAACUGUCGCAAUUUACAUCCCGUCUAAAUGCGGUGUCAGCA-UUGCGACAAAGUAGCACCGAGUUAGACUCGUGU ----..((((((((((((((((((((-.((.......)).)))))))))))..........((((......)))).)))))))-))...........((((.(((.....))))))) ( -45.00, z-score = -3.67, R) >droEre2.scaffold_4845 1256725 113 + 22589142 CAAUAAAGUGUUGAUGCGGCAGUUCU-AGUUUUGCUUAUGAGAACUGUCGCAACUGACAUCCUGUCAAAGUGCUUGGUCGGCAUUUGCAACAUAGUAGCACAUUCCAUUGUUUA--- .......((((((.((((((((((((-.((.......)).))))))))))))..((((.....))))(((((((.....))))))).........)))))).............--- ( -33.50, z-score = -1.79, R) >droYak2.chrU 1961451 111 + 28119190 CAAGAAAACGCUAGUGCGGCAGUUCUCAUUUCUGUUUAUAAGAACUGUCGCAAUGGACAUUCUGUCAAAGUGCUGUGUUGGCACUUGCAACAUUGUAAUAUAUUCCAUUGC------ .........((....))(((((((((.((........)).)))))))))((((((((..........(((((((.....)))))))(((....))).......))))))))------ ( -33.50, z-score = -2.70, R) >droSim1.chrU 2955080 100 + 15797150 ----GAAAUGCUGCUGCGGCAGUUCU-AGGUUUGUUUACCAGAACUGUCGCAAUCGACAUCCUGUCCAC-UGCUACGUCAGCA-UUGCAACAGUGUUGGAGUUAGUG---------- ----..........((((((((((((-.(((......)))))))))))))))...(((.....)))(((-((..((..(((((-(((...))))))))..)))))))---------- ( -38.30, z-score = -3.30, R) >droSec1.super_174 7093 100 - 34763 ----GAAGUGCUGCUGCGGCAGUUCU-AGUUUUGUUUACCAGAACUGUCGCAAUCAACAUCCUGUCCAC-UGCUGCGUCAGCA-UUGCAACAGUGUUGGAGUUAGUG---------- ----.....(((((((((((((((((-.((.......)).)))))))))))..((((((..((((.((.-(((((...)))))-.))..)))))))))))).)))).---------- ( -36.00, z-score = -2.33, R) >droVir3.scaffold_12728 224981 103 + 808307 UUACAAAUUGCCGUUGCAGCAGUUCA-AGUGUCUUGCAUGAGAACUGUCGCAACCAACAU------AAAGUGCUAGAACAGCA-UUGCACCAUAGUGUCACAAUGCUGUGC------ ............(((((.(((((((.-.(((.....)))..))))))).)))))......------...........((((((-((((((....)))...)))))))))..------ ( -32.90, z-score = -1.88, R) >consensus ____AAAGUGCUGCUGCGGCAGUUCU_AGUUUUGUUUACGAGAACUGUCGCAAUCGACAUCCUGUCAAAGUGCUGCGUCAGCA_UUGCAACAUAGUAGCACAUAGCA_UGC______ .........(((((((((((((((((..............)))))))))))......(((.........)))....))))))................................... (-19.03 = -18.92 + -0.11)

| Location | 23,557,000 – 23,557,111 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.59 |
| Shannon entropy | 0.63985 |
| G+C content | 0.45874 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -15.36 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 23557000 111 - 24543557 ACACGAGUCUAACUCGGUGCUACUUUGUCGCAA-UGCUGACACCGCAUUUAGACGGGAUGUAAAUUGCGACAGUUCCCGCAAAAGAAACC-AGAACUGCCGCGCCAGCACUUU---- ...((((.....))))(((((..((((((..((-(((.......)))))..)))))).........(((.((((((..............-.)))))).)))...)))))...---- ( -31.16, z-score = -1.29, R) >droEre2.scaffold_4845 1256725 113 - 22589142 ---UAAACAAUGGAAUGUGCUACUAUGUUGCAAAUGCCGACCAAGCACUUUGACAGGAUGUCAGUUGCGACAGUUCUCAUAAGCAAAACU-AGAACUGCCGCAUCAACACUUUAUUG ---....(((((((.((((((..(((.((((((((((.......))).)))).))).)))..)))((((.(((((((.............-))))))).))))...))).))))))) ( -27.22, z-score = -1.23, R) >droYak2.chrU 1961451 111 - 28119190 ------GCAAUGGAAUAUAUUACAAUGUUGCAAGUGCCAACACAGCACUUUGACAGAAUGUCCAUUGCGACAGUUCUUAUAAACAGAAAUGAGAACUGCCGCACUAGCGUUUUCUUG ------((((((((...((((....(((((.((((((.......))))))))))).))))))))))))..((((((((((........)))))))))).(((....)))........ ( -35.50, z-score = -4.31, R) >droSim1.chrU 2955080 100 - 15797150 ----------CACUAACUCCAACACUGUUGCAA-UGCUGACGUAGCA-GUGGACAGGAUGUCGAUUGCGACAGUUCUGGUAAACAAACCU-AGAACUGCCGCAGCAGCAUUUC---- ----------............(((((((((..-.......))))))-)))....((((((.(.(((((.((((((((((......))).-))))))).)))))).)))))).---- ( -35.80, z-score = -3.29, R) >droSec1.super_174 7093 100 + 34763 ----------CACUAACUCCAACACUGUUGCAA-UGCUGACGCAGCA-GUGGACAGGAUGUUGAUUGCGACAGUUCUGGUAAACAAAACU-AGAACUGCCGCAGCAGCACUUC---- ----------.......(((..(((((((((..-.......))))))-)))....)))(((((.(((((.((((((((((.......)))-))))))).))))))))))....---- ( -42.30, z-score = -5.13, R) >droVir3.scaffold_12728 224981 103 - 808307 ------GCACAGCAUUGUGACACUAUGGUGCAA-UGCUGUUCUAGCACUUU------AUGUUGGUUGCGACAGUUCUCAUGCAAGACACU-UGAACUGCUGCAACGGCAAUUUGUAA ------((((((((((((..(.....)..))))-))))))..(((((....------.)))))((((((.((((((...((.....))..-.)))))).)))))).))......... ( -33.00, z-score = -1.19, R) >consensus ______GCA_AACAAUGUGCUACAAUGUUGCAA_UGCUGACACAGCACUUUGACAGGAUGUCGAUUGCGACAGUUCUCAUAAACAAAACU_AGAACUGCCGCACCAGCACUUU____ .........................(((((....(((.......)))..................((((.((((((................)))))).)))).)))))........ (-15.36 = -14.92 + -0.44)

| Location | 23,557,081 – 23,557,192 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.21 |
| Shannon entropy | 0.65332 |
| G+C content | 0.49717 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 23557081 111 + 24543557 CGACAAAGUAGCACCGAGUUAGACUCGUGUAAGCAAUAAACU---UCUUGGCGGCUUGGCGGGGGGUACCGAAUUGUAUAUAUCCUGUAUGGUGGAUUCGGUAACUCAUGCACA .......((.(((..(((((...(((((.(((((..(((...---..)))...))))))))))....((((((((...(((((...)))))...))))))))))))).))))). ( -29.50, z-score = 0.02, R) >droEre2.scaffold_4845 1256811 114 + 22589142 CAACAUAGUAGCACAUUCCAUUGUUUAUGUGAGCAAUGAACUCUGUCUCGGCGGCUUGGCGGGUGGUACCGAAUCGUAUAUACUCUGUAAUGCGGUAUCGGUAACUCACGCGUA .........(((......((((((((....)))))))).....(((....))))))..(((.(((((((((((((((((.(((...)))))))))).))))).)).))).))). ( -32.60, z-score = -0.53, R) >droSim1.chrU 2955160 103 + 15797150 ----------CAACAGUGUUGGAGUUAGUGGAGCGAUGAACUGCUCCUAGGUGGCUUGGUGGGUGGUACCGAAUUGUGUAUACUCUGUAUAGGCGAUUCGGUAACUCCUCCCC- ----------((((...))))((((((.(((((((......))))))..).))))))((..((.(.(((((((((((.(((((...))))).))))))))))).).))..)).- ( -41.20, z-score = -3.36, R) >droSec1.super_174 7173 101 - 34763 ----------CAACAGUGUUGGAGUUAGUGGGGCGAUGAACUGCUCCUAGGUGGCUUGGCGGGUGGUACCGAAUUGUAUAUACUCUGUAUAGGCGAUUCGGUAACUCGUGC--- ----------((((...))))((((((.(((((((......))))))..).)))))).((((((..(((((((((((.(((((...))))).)))))))))))))))))..--- ( -37.50, z-score = -2.83, R) >droVir3.scaffold_12728 225060 95 + 808307 --------CACCAUAGUGUCA----CAAUGCUGUGCUG--CCGCCGCC--GUUUCUUGGCGGGGUGUACCGAAUCAUUUAUACCCUAUAUGGUAUAUUCAGUAACUCGCGC--- --------((.((((((((..----..)))))))).))--.(((((((--(.....)))))((((.(((.(((.....((((((......))))))))).)))))))))).--- ( -27.40, z-score = -0.74, R) >consensus _________ACCACAGUGUUAGAGUUAGUGGAGCGAUGAACUGCUCCUAGGUGGCUUGGCGGGUGGUACCGAAUUGUAUAUACUCUGUAUAGGCGAUUCGGUAACUCAUGC___ ..........................................................(((.(...(((((((((...(((((...)))))...)))))))))...).)))... (-11.92 = -12.88 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:28 2011