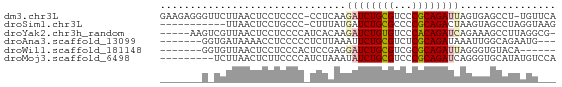
| Sequence ID | dm3.chr3L |
|---|---|
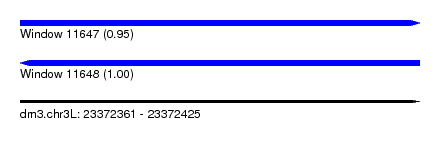
| Location | 23,372,361 – 23,372,425 |
| Length | 64 |
| Max. P | 0.999519 |

| Location | 23,372,361 – 23,372,425 |
|---|---|
| Length | 64 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 60.22 |
| Shannon entropy | 0.74286 |
| G+C content | 0.50599 |
| Mean single sequence MFE | -15.15 |
| Consensus MFE | -7.60 |
| Energy contribution | -7.38 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
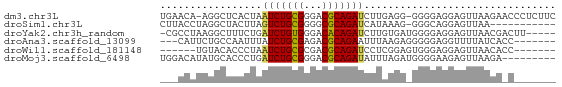
>dm3.chr3L 23372361 64 + 24543557 UGAACA-AGGCUCACUAAUCUGCGGGACGCAGAUCUUGAGG-GGGGAGGAGUUAAGAACCCUCUUC ......-...((((...(((((((...)))))))..)))).-(((((((.((.....))))))))) ( -20.50, z-score = -1.32, R) >droSim1.chr3L 2988948 54 - 22553184 CUUACCUAGGCUACUUAGUCUGCGGGGCGCAGAUCAUAAAG-GGGCAGGAGUUAA----------- ....(((..((..(((.(((((((...)))))))....)))-..)))))......----------- ( -15.10, z-score = -0.84, R) >droYak2.chr3h_random 334991 60 - 911842 -CGCCUAAGGCUUUCUGAUCUGUGGGACACAGAUCUUGUGAUGGGGAGGAGUUAACGACUU----- -..((((..((.....((((((((...))))))))..))..))))(((..(....)..)))----- ( -16.00, z-score = -0.80, R) >droAna3.scaffold_13099 1688301 56 - 3324263 ---CAUUCUGCCAAUUUAUCUGCGAGACGCAGAAUUUAAGAGGGGGAGGUUUUAUCACC------- ---..((((.((..((..((((((...))))))....))..)).))))...........------- ( -13.10, z-score = -1.38, R) >droWil1.scaffold_181148 2074492 53 - 5435427 ------UGUACACCCUAAUCUGCGCGACGCAGAUCCUCGGAGUGGGAGGAGUUAACACC------- ------(((.........((((((...))))))(((((.......)))))....)))..------- ( -13.80, z-score = -0.35, R) >droMoj3.scaffold_6498 3059478 57 + 3408170 UGGACAUAUGCACCCUGAUCUGCGGGACGCAGAUAUUUAGAUGGGGAAGAGUUAAGA--------- ............((((.(((((((...)))))))........))))...........--------- ( -12.40, z-score = -0.95, R) >consensus ___ACAUAGGCUCCCUAAUCUGCGGGACGCAGAUCUUAAGA_GGGGAGGAGUUAACA_C_______ .................(((((((...)))))))................................ ( -7.60 = -7.38 + -0.22)
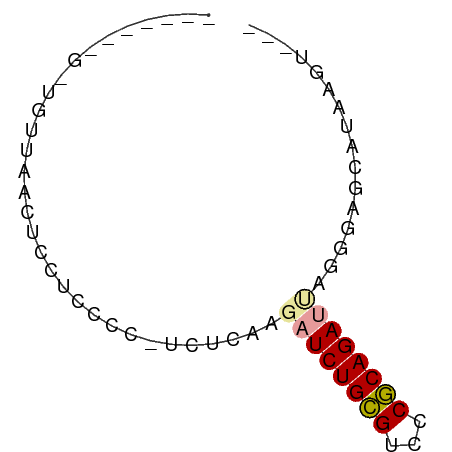
| Location | 23,372,361 – 23,372,425 |
|---|---|
| Length | 64 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 60.22 |
| Shannon entropy | 0.74286 |
| G+C content | 0.50599 |
| Mean single sequence MFE | -14.29 |
| Consensus MFE | -8.11 |
| Energy contribution | -8.42 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
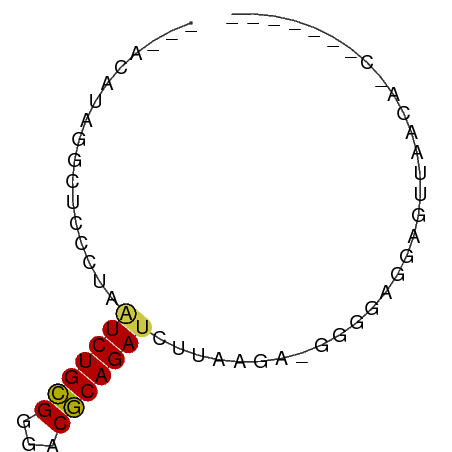
>dm3.chr3L 23372361 64 - 24543557 GAAGAGGGUUCUUAACUCCUCCCC-CCUCAAGAUCUGCGUCCCGCAGAUUAGUGAGCCU-UGUUCA (((.(((((((...(((.......-......((((((((...)))))))))))))))))-).))). ( -20.41, z-score = -2.96, R) >droSim1.chr3L 2988948 54 + 22553184 -----------UUAACUCCUGCCC-CUUUAUGAUCUGCGCCCCGCAGACUAAGUAGCCUAGGUAAG -----------......(((((..-(((...(.((((((...))))))).)))..))..))).... ( -12.30, z-score = -1.81, R) >droYak2.chr3h_random 334991 60 + 911842 -----AAGUCGUUAACUCCUCCCCAUCACAAGAUCUGUGUCCCACAGAUCAGAAAGCCUUAGGCG- -----..........................((((((((...)))))))).....(((...))).- ( -13.50, z-score = -2.80, R) >droAna3.scaffold_13099 1688301 56 + 3324263 -------GGUGAUAAAACCUCCCCCUCUUAAAUUCUGCGUCUCGCAGAUAAAUUGGCAGAAUG--- -------(((......)))((..((........((((((...))))))......))..))...--- ( -11.34, z-score = -1.94, R) >droWil1.scaffold_181148 2074492 53 + 5435427 -------GGUGUUAACUCCUCCCACUCCGAGGAUCUGCGUCGCGCAGAUUAGGGUGUACA------ -------((.(....).))...(((.((...((((((((...)))))))).)))))....------ ( -17.00, z-score = -1.14, R) >droMoj3.scaffold_6498 3059478 57 - 3408170 ---------UCUUAACUCUUCCCCAUCUAAAUAUCUGCGUCCCGCAGAUCAGGGUGCAUAUGUCCA ---------..............(((((....(((((((...)))))))..))))).......... ( -11.20, z-score = -1.90, R) >consensus _______G_UGUUAACUCCUCCCC_UCUCAAGAUCUGCGUCCCGCAGAUUAGGGAGCAUAAGU___ ...............................((((((((...))))))))................ ( -8.11 = -8.42 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:24 2011