
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,339,403 – 23,339,637 |
| Length | 234 |
| Max. P | 0.997981 |

| Location | 23,339,403 – 23,339,512 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 54.23 |
| Shannon entropy | 0.76040 |
| G+C content | 0.51425 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -10.61 |
| Energy contribution | -11.30 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
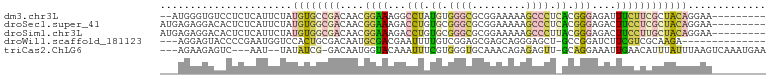
>dm3.chr3L 23339403 109 - 24543557 -----CCUGUCAUUUCAGCCCUA-CAGGCGCCCCCACAUGGGAUGCCUGUGGGAAGCAGGGGUUAAAAGUUUCAAG-AUUCACUUCUACAAGACCACGGGUAAUUUUAAAUAUACA -----(((((.((..(.((((((-(((((((((......))).))))))))))..)).)..)).....((((..((-(......)))...)))).)))))................ ( -32.40, z-score = -1.44, R) >droYak2.chr3L 23921230 106 - 24197627 GCGAAUUUGCGUUCACACCCCCU-CGCGC-UCCGCACAUGGGCGGACUAUGGGAGGCAGGUGUGAAAACUGCAAAACACCUACUCCUACGAG-CGGUGGGAAGCGCACU------- (((..((((((((((((((.(((-(.((.-(((((......)))))...)).))))..))))))))...))))))........((((((...-..))))))..)))...------- ( -44.00, z-score = -2.87, R) >dp4.Unknown_group_112 11972 113 + 17658 GCGAAUUUGCGUUAAUACCGCCG-CGAGC-UCCUCACAUGGGAGGGCUAUGGGAGGUAGGUGUAAAAACUGCCAAGGGCCUACUCCUACGGG-CGAUAGGCAGCGCGCUCCUAACC ........(((((....((.(((-..(((-.((((......))))))).)))..(((((.........)))))..))((((((.((....))-.).)))))))))).......... ( -39.80, z-score = -0.03, R) >triCas2.ChLG6 13063411 112 - 13544221 ---UAUAUGGAAGUCUAUUCCUGCCAAGUCUCCAUAUACCGGGGGAAUUUGGGAAGCGGGGGUUAAAACAUUUAAGCAUCAUUUUAAACGUG-UUGUGGGACUCUCGUUAUUAACA ---.....((((.....))))..((((((((((........)))).))))))..((((((((((..((((((((((......)))))..)))-))....))))))))))....... ( -30.20, z-score = -1.32, R) >consensus ___AAUUUGCGAUUACACCCCCG_CAAGC_UCCACACAUGGGAGGACUAUGGGAAGCAGGGGUUAAAACUGCCAAGCACCUACUCCUACGAG_CGAUGGGAAGCGCGCUAUUAACA ........(((((.....((((.....((.(((...(((((.....)))))))).)).))))......................(((((......))))).))))).......... (-10.61 = -11.30 + 0.69)


| Location | 23,339,442 – 23,339,543 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 54.19 |
| Shannon entropy | 0.75753 |
| G+C content | 0.49140 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -14.06 |
| Energy contribution | -15.12 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.997981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 23339442 101 - 24543557 -----CAAGUUACCUCGUCGAGCCA----UCAAAGCCUAUCCUGUCAUUUCAGCCCUACAGGCGCCCCCACAUGGGAUGCCUGUGGGAAGCAGGGGUUAAAAGUUUCAAG -----......(((((...((....----))...((.....(((......))).(((((((((((((......))).))))))))))..)).)))))............. ( -31.80, z-score = -1.48, R) >droYak2.chr3L 23921262 104 - 24197627 -----CAAUUCGCGAGUUCGCAUCAAAAAGCGGAUGCGAAUUUGCGUUCACACCCCCUCGCGC-UCCGCACAUGGGCGGACUAUGGGAGGCAGGUGUGAAAACUGCAAAA -----.....((((((((((((((........))))))))))))))((((((((.((((.((.-(((((......)))))...)).))))..)))))))).......... ( -49.50, z-score = -5.17, R) >dp4.Unknown_group_112 12011 104 + 17658 -----CAAUUCGCGACUUCGCGUCAAAAAACGGAUGCGAAUUUGCGUUAAUACCGCCGCGAGC-UCCUCACAUGGGAGGGCUAUGGGAGGUAGGUGUAAAAACUGCCAAG -----.....(((((.((((((((........)))))))).)))))....((((.((.(((((-.((((......))))))).)))).))))(((((....)).)))... ( -38.00, z-score = -2.83, R) >triCas2.ChLG6 13063450 110 - 13544221 AAAAGAAAGUAGUCAAAAGAUUAAAUAAUGAAGGUAUUAUAUGGAAGUCUAUUCCUGCCAAGUCUCCAUAUACCGGGGGAAUUUGGGAAGCGGGGGUUAAAACAUUUAAG .........(((((....))))).............(((.(((.......(((((((((((((((((........)))).)))))....)))))))).....))).))). ( -20.30, z-score = 0.19, R) >consensus _____CAAGUCGCCACUUCGCGUCAAAAAGCAGAUGCGAAUUUGCGUUCACACCCCCACAAGC_UCCACACAUGGGAGGACUAUGGGAAGCAGGGGUUAAAACUGCCAAG ...........((((.((((((((........)))))))).))))........((((....((.(((...(((((.....)))))))).)).)))).............. (-14.06 = -15.12 + 1.06)



| Location | 23,339,446 – 23,339,547 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 54.77 |
| Shannon entropy | 0.72356 |
| G+C content | 0.50109 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -12.19 |
| Energy contribution | -14.25 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 23339446 101 - 24543557 --------GGCACAAGUUACCUCGUCGAGCCAUCAA----AGCCUAUCCUGUCAUUUCAGCCCUACAGGCGCCCCCACAUGGGAUGCCUGUGGGAAGCAGGGGUUAAAAGUUU --------(((((.((....)).))...))).....----(((((...(((......))).(((((((((((((......))).))))))))))......)))))........ ( -32.30, z-score = -0.72, R) >droYak2.chr3L 23921266 104 - 24197627 --------GACGCAAUUCGCGAGUUCGCAUCAAAAAGCGGAUGCGAAUUUGCGUUCACACCCCCUCGCGC-UCCGCACAUGGGCGGACUAUGGGAGGCAGGUGUGAAAACUGC --------...(((...((((((((((((((........))))))))))))))((((((((.((((.((.-(((((......)))))...)).))))..))))))))...))) ( -51.10, z-score = -5.11, R) >dp4.Unknown_group_112 12015 104 + 17658 --------GACACAAUUCGCGACUUCGCGUCAAAAAACGGAUGCGAAUUUGCGUUAAUACCGCCGCGAGC-UCCUCACAUGGGAGGGCUAUGGGAGGUAGGUGUAAAAACUGC --------.((((....(((((.((((((((........)))))))).)))))....((((.((.(((((-.((((......))))))).)))).)))).))))......... ( -39.80, z-score = -3.62, R) >triCas2.ChLG6 13063454 110 - 13544221 UUUGAAAAGAAAGUAGUCAAAAGAUUAAAUAAUGAA---GGUAUUAUAUGGAAGUCUAUUCCUGCCAAGUCUCCAUAUACCGGGGGAAUUUGGGAAGCGGGGGUUAAAACAUU .......(((...(((((....))))).((((((..---..)))))).......)))(((((((((((((((((........)))).)))))....))))))))......... ( -19.70, z-score = 0.63, R) >consensus ________GACACAAGUCACGACUUCGAAUCAAAAA___GAUGCGAAUUUGCGUUCACACCCCCACAAGC_UCCACACAUGGGAGGACUAUGGGAAGCAGGGGUUAAAACUGC ..................((((.((((((((........)))))))).))))........((((....((.(((...(((((.....)))))))).)).)))).......... (-12.19 = -14.25 + 2.06)


| Location | 23,339,547 – 23,339,637 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 55.17 |
| Shannon entropy | 0.76839 |
| G+C content | 0.50785 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -7.14 |
| Energy contribution | -9.14 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 23339547 90 - 24543557 --AUGGGUGUCCUCUCAUUCUAUGUGCCGACAACGGAAAGGCCUAUGUGGGCGCGGAAAAAGCCCUCACGGGAGAUUUCUUCGCUACAGGAA--------- --(((((......)))))(((.((((.(((....(((((..((..((.((((.........)))).))..))...)))))))).))))))).--------- ( -26.60, z-score = 0.04, R) >droSec1.super_41 221023 92 - 296808 AUGAGAGGACACUCUCAUUCUAUGUGGCGACAACGGAAAGACCUGUGCGGGCGCGGAAAAAGCCCUCACGGGAGACUUCCUCGCUACAGGAA--------- (((((((....)))))))(((.((((((((....((((...((((((.((((.........)))).))))))....))))))))))))))).--------- ( -42.00, z-score = -4.94, R) >droSim1.chr3L 22460309 92 - 22553184 AUGAGAGGACACUCUCAUUCUAUGUGGCGACAACGGAAAGACCUGUGCGGGCGCGGAAAAAGCCCUUACGGGAGACUUCCUUGCUACAGGAA--------- (((((((....)))))))(((.((((((((....((((...((((((.((((.........)))).))))))....))))))))))))))).--------- ( -37.70, z-score = -4.19, R) >droWil1.scaffold_181123 1341468 83 - 1459946 ---AGGAGUACCCCGAAUGGUCCACUGCGACAAUGCGACGAAUUUUGUCGGAGCGAGCAGGGAGCU-GCCGGAUCUUCGUCGCAAGA-------------- ---.((.....))((((.(((((.((.((((((...........)))))).)).(.((((....))-))))))))))))((....))-------------- ( -27.70, z-score = -0.46, R) >triCas2.ChLG6 13063564 91 - 13544221 ---AGAAGAGUC---AAU--UAUAUCG-GACAAUGGUACAAAUUUCGUGGGUGCAAACAGAGAGUU-GCAGGAAAUUGAACAUUUAUUUAAGUCAAAUGAA ---.(((...((---(((--(.(((((-.....)))))(((.((((.((........)))))).))-).....))))))...)))................ ( -9.70, z-score = 1.50, R) >consensus ___AGAGGACACUCUCAUUCUAUGUGGCGACAACGGAAAGACCUGUGUGGGCGCGGAAAAAGCCCU_ACGGGAGAUUUCCUCGCUACAGGAA_________ ......................((((((((....((((...(((.((.((((.........)))).)).)))....))))))))))))............. ( -7.14 = -9.14 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:22 2011