
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,231,303 – 23,231,354 |
| Length | 51 |
| Max. P | 0.994031 |

| Location | 23,231,303 – 23,231,354 |
|---|---|
| Length | 51 |
| Sequences | 3 |
| Columns | 53 |
| Reading direction | forward |
| Mean pairwise identity | 56.69 |
| Shannon entropy | 0.62825 |
| G+C content | 0.32889 |
| Mean single sequence MFE | -10.70 |
| Consensus MFE | -9.82 |
| Energy contribution | -8.27 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.994031 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
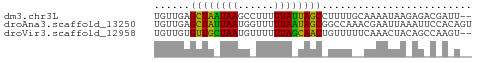
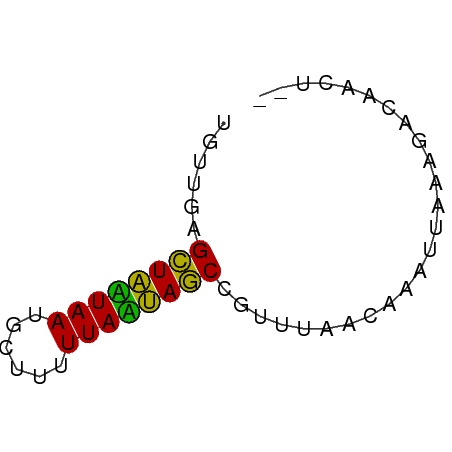
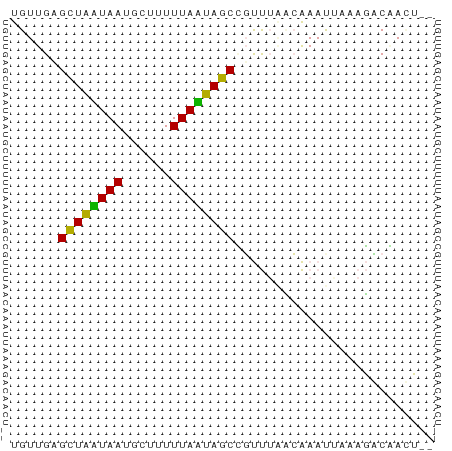
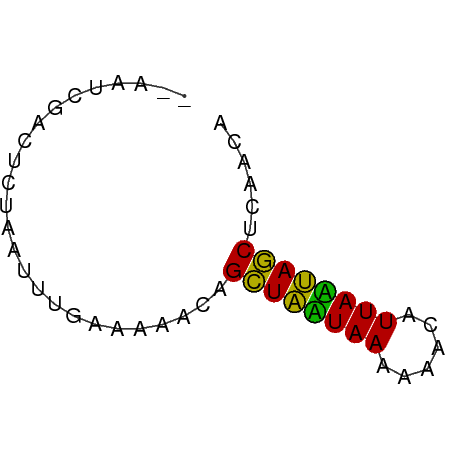
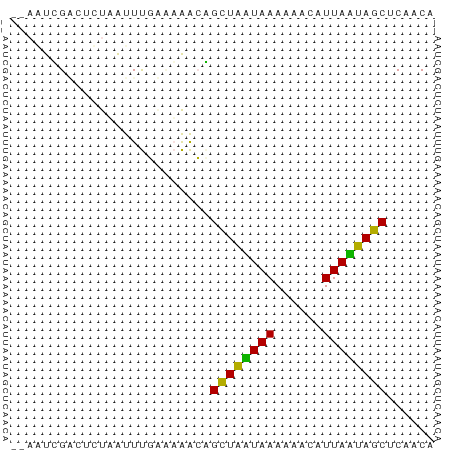
>dm3.chr3L 23231303 51 + 24543557 UGUUGAGCUAAUAAGCCUUUUUAUUAGCCUUUUGCAAAAUAAGAGACGAUU-- .((((.(((((((((....)))))))))((((((.....)))))).)))).-- ( -9.90, z-score = -1.77, R) >droAna3.scaffold_13250 1155462 53 + 3535662 UGUUGAGCUAUUAAUGGUUUUUAAUAGCGGCCAAACGAAUUAAAUUCCACAGU (((.((((((((((......))))))))(......).........)).))).. ( -9.40, z-score = -1.06, R) >droVir3.scaffold_12958 668084 51 - 3547706 UGUUGUGUUGCUAAUGUUUUUUAGCAACUGUUUUUCAAACUACAGCCAAGU-- .(((((((((((((......)))))))).(((.....))).))))).....-- ( -12.80, z-score = -1.98, R) >consensus UGUUGAGCUAAUAAUGCUUUUUAAUAGCCGUUUAACAAAUUAAAGACAACU__ ......((((((((......))))))))......................... ( -9.82 = -8.27 + -1.55)
| Location | 23,231,303 – 23,231,354 |
|---|---|
| Length | 51 |
| Sequences | 3 |
| Columns | 53 |
| Reading direction | reverse |
| Mean pairwise identity | 56.69 |
| Shannon entropy | 0.62825 |
| G+C content | 0.32889 |
| Mean single sequence MFE | -9.07 |
| Consensus MFE | -9.69 |
| Energy contribution | -8.13 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.22 |
| Structure conservation index | 1.07 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963706 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
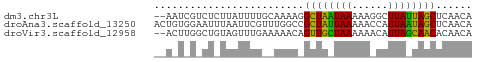
>dm3.chr3L 23231303 51 - 24543557 --AAUCGUCUCUUAUUUUGCAAAAGGCUAAUAAAAAGGCUUAUUAGCUCAACA --......................(((((((((......)))))))))..... ( -8.10, z-score = -1.33, R) >droAna3.scaffold_13250 1155462 53 - 3535662 ACUGUGGAAUUUAAUUCGUUUGGCCGCUAUUAAAAACCAUUAAUAGCUCAACA ...((.((((.......)))).)).((((((((......))))))))...... ( -8.90, z-score = -1.25, R) >droVir3.scaffold_12958 668084 51 + 3547706 --ACUUGGCUGUAGUUUGAAAAACAGUUGCUAAAAAACAUUAGCAACACAACA --.....(.(((.(((.....))).((((((((......))))))))))).). ( -10.20, z-score = -1.09, R) >consensus __AAUCGACUCUAAUUUGAAAAACAGCUAAUAAAAAACAUUAAUAGCUCAACA .........................((((((((......))))))))...... ( -9.69 = -8.13 + -1.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:15 2011