
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,138,049 – 23,138,126 |
| Length | 77 |
| Max. P | 0.835168 |

| Location | 23,138,049 – 23,138,126 |
|---|---|
| Length | 77 |
| Sequences | 4 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 67.47 |
| Shannon entropy | 0.54861 |
| G+C content | 0.39666 |
| Mean single sequence MFE | -13.70 |
| Consensus MFE | -10.72 |
| Energy contribution | -10.40 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
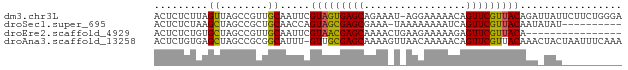
>dm3.chr3L 23138049 77 + 24543557 ACUCUCUUAGUUAGCCGUUGCAAUUCGUAGUGAGCAGAAAU-AGGAAAAACAGUUCGUUACAGAUUAUUCUUCUGGGA ....((((((...((....))(((..(((((((((......-..........)))))))))..)))......)))))) ( -11.39, z-score = 1.24, R) >droSec1.super_695 2973 67 + 7776 ACUCUCUAAGCUAGCCGCUGCAACCAGUAGCGAGCGAAA-UAAAAAAAAUCAGUUCGUUACAAUAUAU---------- ........(((.....))).......(((((((((....-............))))))))).......---------- ( -13.39, z-score = -2.02, R) >droEre2.scaffold_4929 3191150 62 - 26641161 ACUCUCUGUGCUAGCCGUUGCAAUUCGUAACGAGCAAAACUGAAGAAAAAGAGUUCGUUACA---------------- ........(((........)))....(((((((((....((........)).))))))))).---------------- ( -13.50, z-score = -0.75, R) >droAna3.scaffold_13258 104985 77 + 1584923 ACUCUGUGAGCUAGCCGCGGCAUUU-GUUGCGAGCAAAAGUUAACAAAAACAGUUCGUUACAAACUACUAAUUUCAAA .......(((((.((((((((....-)))))).))....(((......))))))))...................... ( -16.50, z-score = -0.79, R) >consensus ACUCUCUGAGCUAGCCGCUGCAAUUCGUAGCGAGCAAAA_U_AAGAAAAACAGUUCGUUACAAA_UAU__________ .........((........)).....(((((((((.................)))))))))................. (-10.72 = -10.40 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:12 2011