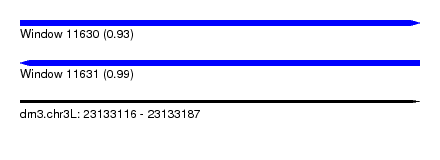
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,133,116 – 23,133,187 |
| Length | 71 |
| Max. P | 0.993676 |

| Location | 23,133,116 – 23,133,187 |
|---|---|
| Length | 71 |
| Sequences | 5 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.53272 |
| G+C content | 0.41287 |
| Mean single sequence MFE | -15.58 |
| Consensus MFE | -12.26 |
| Energy contribution | -11.82 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
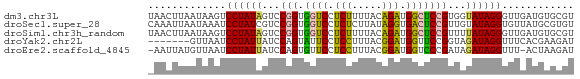
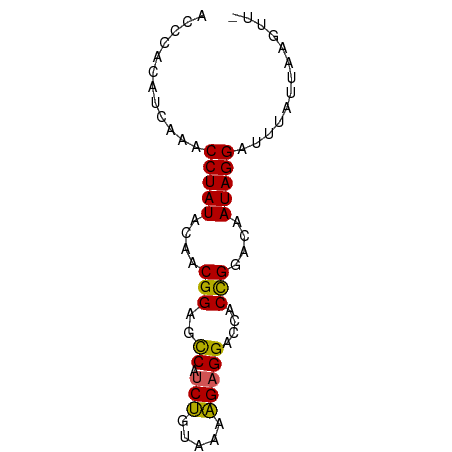
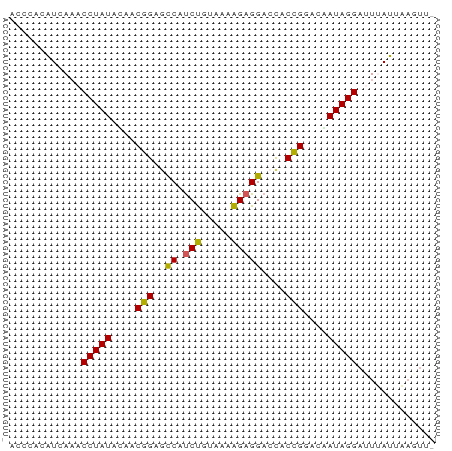
>dm3.chr3L 23133116 71 + 24543557 UAACUUAAUAAGUCCUAUAGUCCGGUGGUCCUCUUUUACAGAUGGCUCCGUGGUAUAGGGUUGAUGUGCGU ..........(..((((((.(.(((.((((.(((.....))).))))))).).))))))..)......... ( -18.40, z-score = -0.79, R) >droSec1.super_28 416 71 - 873875 CAAAUUAAUAAAUCCUAUCGUCCGGUGGUCCUCUCUUAUAGGUGACUCCGUUGUAUAGGUGUUAUGCGUGU .......((((..(((((((..(((.((((.(((.....))).))))))).)).)))))..))))...... ( -14.40, z-score = -1.21, R) >droSim1.chr3h_random 13375 71 - 1452968 UAACUUAAUAAGUCCUAUAGUCCGGUGGUCCUCUUUUACAGAUGGCUCCGUUUUAUAGGGUUGAUGUGCGU ..........(..(((((((..(((.((((.(((.....))).)))))))..)))))))..)......... ( -19.90, z-score = -2.03, R) >droYak2.chr2L 73137 64 - 22324452 -------GUUAAUCCUAUUAUCCAGUAUUCCUCCUUUACGGAUGGUUCCGGUAGAUAGGUUUCACGAAGAU -------((.((.((((((..((.(....(((((.....))).))..).))..)))))).)).))...... ( -13.50, z-score = -0.53, R) >droEre2.scaffold_4845 953297 69 + 22589142 -AAUUAUGUUAAUCCUAUUAUCCAGUGUUCCUCCUUUACGGAUGGUCCCGAUAGAUAGGUUU-ACUAAGAU -........(((.(((((((((..(.(..(((((.....))).))..))))).)))))).))-)....... ( -11.70, z-score = 0.06, R) >consensus _AACUUAAUAAAUCCUAUAGUCCGGUGGUCCUCUUUUACAGAUGGCUCCGUUAUAUAGGUUUGAUGAGCGU .............((((((...(((.((((.(((.....))).)))))))...))))))............ (-12.26 = -11.82 + -0.44)

| Location | 23,133,116 – 23,133,187 |
|---|---|
| Length | 71 |
| Sequences | 5 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.53272 |
| G+C content | 0.41287 |
| Mean single sequence MFE | -14.72 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.04 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
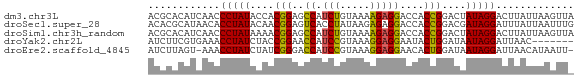
>dm3.chr3L 23133116 71 - 24543557 ACGCACAUCAACCCUAUACCACGGAGCCAUCUGUAAAAGAGGACCACCGGACUAUAGGACUUAUUAAGUUA ............((((((...(((..((.(((.....)))))....)))...))))))............. ( -14.60, z-score = -1.46, R) >droSec1.super_28 416 71 + 873875 ACACGCAUAACACCUAUACAACGGAGUCACCUAUAAGAGAGGACCACCGGACGAUAGGAUUUAUUAAUUUG ............(((((....(((.(((..((.....))..)))..)))....)))))............. ( -14.90, z-score = -3.42, R) >droSim1.chr3h_random 13375 71 + 1452968 ACGCACAUCAACCCUAUAAAACGGAGCCAUCUGUAAAAGAGGACCACCGGACUAUAGGACUUAUUAAGUUA ............((((((...(((..((.(((.....)))))....)))...))))))............. ( -14.60, z-score = -1.65, R) >droYak2.chr2L 73137 64 + 22324452 AUCUUCGUGAAACCUAUCUACCGGAACCAUCCGUAAAGGAGGAAUACUGGAUAAUAGGAUUAAC------- ............(((((...((((..((.(((.....)))))....))))...)))))......------- ( -14.40, z-score = -0.98, R) >droEre2.scaffold_4845 953297 69 - 22589142 AUCUUAGU-AAACCUAUCUAUCGGGACCAUCCGUAAAGGAGGAACACUGGAUAAUAGGAUUAACAUAAUU- ......((-.(((((((.((((.((.((.(((.....)))))....)).))))))))).)).))......- ( -15.10, z-score = -1.28, R) >consensus ACCCACAUCAAACCUAUACAACGGAGCCAUCUGUAAAAGAGGACCACCGGACAAUAGGAUUUAUUAAGUU_ ............(((((....(((..((.(((.....)))))....)))....)))))............. (-11.72 = -11.04 + -0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:11 2011