| Sequence ID | dm3.chr3L |
|---|---|
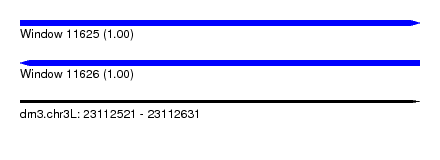
| Location | 23,112,521 – 23,112,631 |
| Length | 110 |
| Max. P | 0.999452 |

| Location | 23,112,521 – 23,112,631 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 52.14 |
| Shannon entropy | 0.76593 |
| G+C content | 0.44120 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -13.34 |
| Energy contribution | -16.27 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
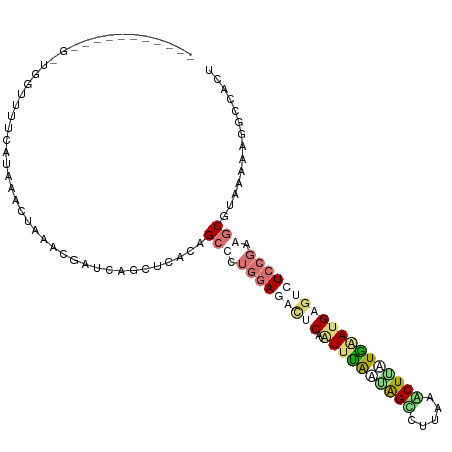
>dm3.chr3L 23112521 110 + 24543557 GCGUAGCUAGAGAUAGUUUUCGUCGAGCAGAUGAUCUUCUUUCCGCGCUUAAUUCACAGUUUAAUGGCCUUAAGCUAAUAGAGGAAUCUCCGAAGCGCAAAGAAGCCGCU (((.(((((....))))).(((((.....))))).(((((((..((((((.........((((.((((.....)))).))))((.....)).))))))))))))).))). ( -29.50, z-score = -0.25, R) >droPer1.super_76 130794 89 - 268861 ---------------------AAAAGCAAUACCAUCGGCUCUUAGCCAUGGAGUCUCAAUAUGGCAGUCUGCAACUUCUGAAUGAGUCUCCGAGGCGUAAAAAGGUCACU ---------------------...............(((.(((.(((.(((((.((((...(((.(((.....))).)))..)))).))))).))).....))))))... ( -22.90, z-score = -1.12, R) >droWil1.scaffold_181148 2801111 101 - 5435427 ---------GGGCAGGUUUUAAUAAACUCCAAGCGGAGUUUACUGCGGUGGAGACUCAGUUCAGAAGUUUAUCGCUUAUGAAUGAGUCUCCGAAACGUAAAAGGUCCAGU ---------((((.........((((((((....)))))))).((((.((((((((((.((((.((((.....)))).))))))))))))))...))))....))))... ( -41.90, z-score = -5.92, R) >droVir3.scaffold_12958 1203962 99 - 3547706 -----------GUUGGUUUUCGUAAACUUAACGAUCAACUCACAGCCCUGGACAUCCAAUUUAAUAGCCUUAAACUGUUGAAUGAGUCUCCUGGGCCUAAAAAGUCCAAU -----------((.((((.((((.......))))..)))).)).((((.(((.(..((.((((((((.......))))))))))..).))).)))).............. ( -24.20, z-score = -2.47, R) >consensus ___________G_UGGUUUUCAUAAACUAAACGAUCAGCUCACAGCCCUGGAGACUCAAUUUAAUAGCCUUAAACUUAUGAAUGAGUCUCCGAAGCGUAAAAAGGCCACU ............................................(((.(((((((((.((((((((((.....))))))))))))))))))).))).............. (-13.34 = -16.27 + 2.94)

| Location | 23,112,521 – 23,112,631 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 52.14 |
| Shannon entropy | 0.76593 |
| G+C content | 0.44120 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -12.68 |
| Energy contribution | -17.30 |
| Covariance contribution | 4.62 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
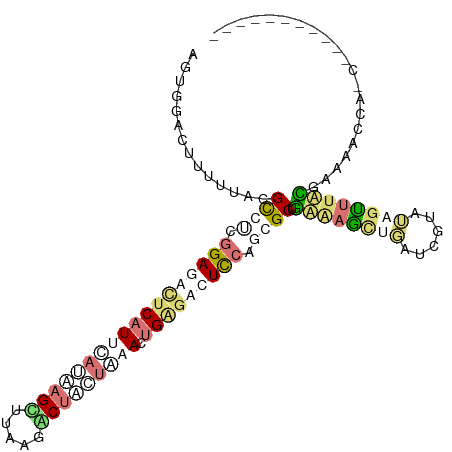
>dm3.chr3L 23112521 110 - 24543557 AGCGGCUUCUUUGCGCUUCGGAGAUUCCUCUAUUAGCUUAAGGCCAUUAAACUGUGAAUUAAGCGCGGAAAGAAGAUCAUCUGCUCGACGAAAACUAUCUCUAGCUACGC .(((.(((((((.(((...((((....))))....((((((..(((......)).)..))))))))).))))))).......(((.((.((......)))).)))..))) ( -31.60, z-score = -1.12, R) >droPer1.super_76 130794 89 + 268861 AGUGACCUUUUUACGCCUCGGAGACUCAUUCAGAAGUUGCAGACUGCCAUAUUGAGACUCCAUGGCUAAGAGCCGAUGGUAUUGCUUUU--------------------- .((((.....))))(((((((((.((((......((((...)))).......)))).))))..(((.....))))).))).........--------------------- ( -22.02, z-score = -0.70, R) >droWil1.scaffold_181148 2801111 101 + 5435427 ACUGGACCUUUUACGUUUCGGAGACUCAUUCAUAAGCGAUAAACUUCUGAACUGAGUCUCCACCGCAGUAAACUCCGCUUGGAGUUUAUUAAAACCUGCCC--------- ...((..............(((((((((((((.(((.......))).)))).))))))))).....((((((((((....))))))))))....)).....--------- ( -32.80, z-score = -4.97, R) >droVir3.scaffold_12958 1203962 99 + 3547706 AUUGGACUUUUUAGGCCCAGGAGACUCAUUCAACAGUUUAAGGCUAUUAAAUUGGAUGUCCAGGGCUGUGAGUUGAUCGUUAAGUUUACGAAAACCAAC----------- .((((((((....(((((.(((....((((((...((((((.....))))))))))))))).)))))..))))...((((.......))))...)))).----------- ( -26.50, z-score = -1.70, R) >consensus AGUGGACUUUUUACGCCUCGGAGACUCAUUCAUAAGCUUAAGACUACUAAACUGAGACUCCAGCGCUGAAAGCUGAUCGUAUAGUUUACGAAAACCA_C___________ ..............((((.((((((((((((((((((.....))))))))).))))))))).)))).((((((((......))))))))..................... (-12.68 = -17.30 + 4.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:07 2011