| Sequence ID | dm3.chr3L |
|---|---|
| Location | 23,041,667 – 23,041,744 |
| Length | 77 |
| Max. P | 0.946321 |

| Location | 23,041,667 – 23,041,744 |
|---|---|
| Length | 77 |
| Sequences | 4 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 60.68 |
| Shannon entropy | 0.64759 |
| G+C content | 0.53011 |
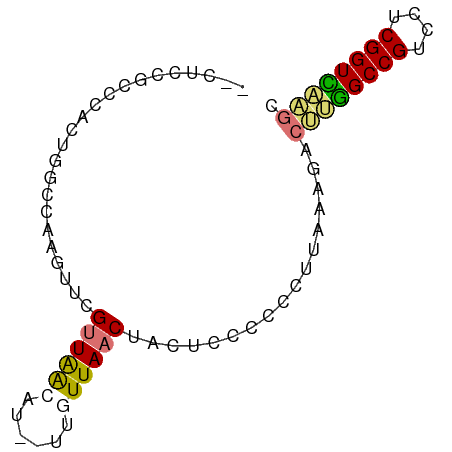
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -10.61 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
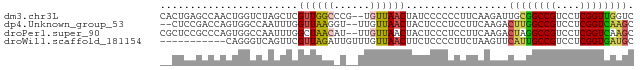
>dm3.chr3L 23041667 77 - 24543557 CACUGAGCCAACUGGUCUAGCUCGUUGGCCCG--UGUUAACUAUCCCCCCUUCAAGAUUGCGGCCGUCCUCGGUUGGUC (((.(.((((((.((.....)).))))))).)--))........................((((((....))))))... ( -21.00, z-score = -0.48, R) >dp4.Unknown_group_53 32161 75 - 37457 --CUCCGACCAGUGGCCAAUUUGGUUAAGGU--UUGUUAACUACUCCCUCCUUCAAGACUUGGCCGUCCUCGGUCAAGC --....((((((((((((((((((...(((.--..((.....))..)))...)))))..)))))))..)).)))).... ( -21.20, z-score = -1.58, R) >droPer1.super_90 61290 77 - 211371 CGCUCCGCCCAGUGGCCAAUUUGGCUAACAU--UUGUUAACUACUCCCUCCUUCAAGACUAGGCCGUCCUCGGUCAAGC .(((..(((.(((((((....(((.((((..--..)))).)))....((......))....)))))..)).)))..))) ( -16.80, z-score = -0.40, R) >droWil1.scaffold_181154 1378002 68 - 4610121 -----------CAGGGUCAGUUCGUUAGAUUGUUUGUUAACUUCUCCCCUUCUAAGUUCAUUGCCGUCCUCGGUGAUGC -----------.((((..((...(((((........)))))..)).))))........(((..(((....)))..))). ( -15.30, z-score = -1.67, R) >consensus __CUCCGCCCACUGGCCAAGUUCGUUAACAU__UUGUUAACUACUCCCCCCUUAAAGACUUGGCCGUCCUCGGUCAAGC .......................((((((......)))))).................((((((((....)))))))). (-10.61 = -10.67 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:46:06 2011