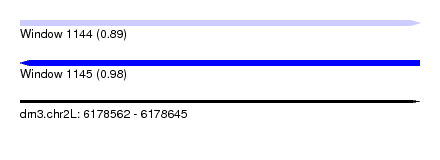
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,178,562 – 6,178,645 |
| Length | 83 |
| Max. P | 0.977605 |

| Location | 6,178,562 – 6,178,645 |
|---|---|
| Length | 83 |
| Sequences | 7 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 67.99 |
| Shannon entropy | 0.57473 |
| G+C content | 0.54253 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
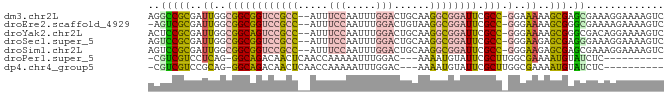
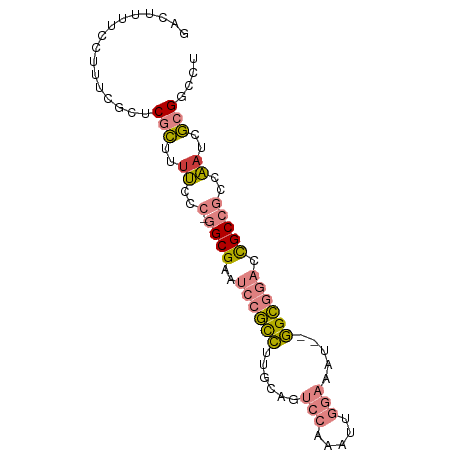
>dm3.chr2L 6178562 83 + 23011544 AGGCCGCGAUUGGCGGCGGUCCGCC--AUUUCCAAUUUGGACUGCAAGGCGGAUUCGCC-GGAAAAAGCGAGCGAAAGGAAAAGUC ..(((((..((..((((((((((((--...(((.....)))......)))))).)))))-)..))..))).))............. ( -32.30, z-score = -1.42, R) >droEre2.scaffold_4929 15106245 82 + 26641161 -AGUCGCGAUUGGCGGCGGUCCGCC--AUUUCCAAUUUGGACUGUAAGGCGGAUUCGCC-GGGAAAAGCGGGCGAAAAGAAAAGUC -.(((((.....))))).(((((((--...(((.....)))......)))))))(((((-.(......).)))))........... ( -30.60, z-score = -1.56, R) >droYak2.chr2L 15593150 83 - 22324452 ACUCCGCGAUUGGCGGCAGUCCGCC--AUUUCCAAUUUGGACUGCAAGGCGGAUUCGCC-GGGAAAAGCGGGCGACAGGAAAAGUC ..(((((..((..((((((((((((--...(((.....)))......)))))))).)))-)..))..))))).(((.......))) ( -31.50, z-score = -1.28, R) >droSec1.super_5 4249868 83 + 5866729 AGUCCGCGAUUGGCGGCGGUCCGCC--AUUUCCAAUUUGGACUGCAAGGCGGAUUCGCC-GGGAAGAGCGAGGGAAAGGAAAAGUC ..(((((..((..((((((((((((--...(((.....)))......)))))).)))))-)..))..))).))............. ( -30.00, z-score = -0.77, R) >droSim1.chr2L 5971459 83 + 22036055 AGUCCGCGAUUGGCGGCGGUCCGCC--AUUUCCAAUUUGGACUGCAAGGCGGAUUCGCC-GGGAAGAGCGAGCGAAAGGAAAAGUC .(..(((..((..((((((((((((--...(((.....)))......)))))).)))))-)..))..)))..)............. ( -32.40, z-score = -1.36, R) >droPer1.super_5 38919 71 - 6813705 -CGUCGUCCUCAG-GGCAGACAACUCAACCAAAAAUUUGGAC---AAAAUGUAUUCGCUUGGCGAAAAUGUAUCUC---------- -.(((((((...)-))).))).......((((....))))..---...((((.(((((...))))).)))).....---------- ( -14.10, z-score = -0.94, R) >dp4.chr4_group5 39529 71 - 2436548 -CGUCGUCCGCAG-GGCAGACAACUCAACCAAAAAUUUGGAC---AAAAUGUAUUCGCUUGGCGAAAAUGUAUCUC---------- -.(((((((...)-))).))).......((((....))))..---...((((.(((((...))))).)))).....---------- ( -14.10, z-score = -0.50, R) >consensus ACGCCGCGAUUGGCGGCGGUCCGCC__AUUUCCAAUUUGGACUGCAAGGCGGAUUCGCC_GGGAAAAGCGAGCGAAAGGAAAAGUC ..(((((.......(((((((((((.....(((.....)))......)))))))).)))........))).))............. (-15.30 = -15.02 + -0.28)

| Location | 6,178,562 – 6,178,645 |
|---|---|
| Length | 83 |
| Sequences | 7 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 67.99 |
| Shannon entropy | 0.57473 |
| G+C content | 0.54253 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -13.06 |
| Energy contribution | -13.41 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
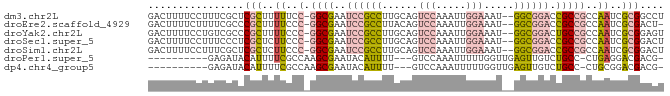
>dm3.chr2L 6178562 83 - 23011544 GACUUUUCCUUUCGCUCGCUUUUUCC-GGCGAAUCCGCCUUGCAGUCCAAAUUGGAAAU--GGCGGACCGCCGCCAAUCGCGGCCU .............((.(((..((..(-((((..((((((......(((.....)))...--)))))).)))))..))..))))).. ( -30.10, z-score = -1.98, R) >droEre2.scaffold_4929 15106245 82 - 26641161 GACUUUUCUUUUCGCCCGCUUUUCCC-GGCGAAUCCGCCUUACAGUCCAAAUUGGAAAU--GGCGGACCGCCGCCAAUCGCGACU- ................(((..((..(-((((..((((((......(((.....)))...--)))))).)))))..))..)))...- ( -27.50, z-score = -2.62, R) >droYak2.chr2L 15593150 83 + 22324452 GACUUUUCCUGUCGCCCGCUUUUCCC-GGCGAAUCCGCCUUGCAGUCCAAAUUGGAAAU--GGCGGACUGCCGCCAAUCGCGGAGU (((.......)))((((((..((..(-((((..((((((......(((.....)))...--)))))).)))))..))..)))).)) ( -31.90, z-score = -2.29, R) >droSec1.super_5 4249868 83 - 5866729 GACUUUUCCUUUCCCUCGCUCUUCCC-GGCGAAUCCGCCUUGCAGUCCAAAUUGGAAAU--GGCGGACCGCCGCCAAUCGCGGACU ...............((((......(-((((..((((((......(((.....)))...--)))))).)))))......))))... ( -27.30, z-score = -1.87, R) >droSim1.chr2L 5971459 83 - 22036055 GACUUUUCCUUUCGCUCGCUCUUCCC-GGCGAAUCCGCCUUGCAGUCCAAAUUGGAAAU--GGCGGACCGCCGCCAAUCGCGGACU .............(..(((......(-((((..((((((......(((.....)))...--)))))).)))))......)))..). ( -28.40, z-score = -1.72, R) >droPer1.super_5 38919 71 + 6813705 ----------GAGAUACAUUUUCGCCAAGCGAAUACAUUUU---GUCCAAAUUUUUGGUUGAGUUGUCUGCC-CUGAGGACGACG- ----------((((.....))))((((((((((.....)))---))........)))))...(((((((...-....))))))).- ( -14.50, z-score = -0.54, R) >dp4.chr4_group5 39529 71 + 2436548 ----------GAGAUACAUUUUCGCCAAGCGAAUACAUUUU---GUCCAAAUUUUUGGUUGAGUUGUCUGCC-CUGCGGACGACG- ----------((((.....))))((((((((((.....)))---))........)))))...(((((((((.-..))))))))).- ( -18.70, z-score = -1.90, R) >consensus GACUUUUCCUUUCGCUCGCUUUUCCC_GGCGAAUCCGCCUUGCAGUCCAAAUUGGAAAU__GGCGGACCGCCGCCAAUCGCGGCCU ................(((..((....((((..((((((......(((.....))).....)))))).))))...))..))).... (-13.06 = -13.41 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:37 2011