| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,883,083 – 22,883,180 |
| Length | 97 |
| Max. P | 0.761438 |

| Location | 22,883,083 – 22,883,180 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.27 |
| Shannon entropy | 0.60705 |
| G+C content | 0.51392 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22883083 97 - 24543557 UUCGAGGGU-GCCAAGCGAAAGUCGGCC--UUGAUUCUUCCGAGG-AUCGA---UAA-GCUUUCGCUGGUGC-CGUUUCAAAGUGCGAGUACUCACAAAC-CGUCAG ...(((((.-.(((.(((((((((((((--(((.......)))))-.))).---...-)))))))))))..)-)((........)).....)))......-...... ( -33.80, z-score = -1.98, R) >droPer1.super_9 2339812 106 + 3637205 UUCAAGGACAGCCCAGCGAAAGUGGGCUUAUUGAUUCUUUCUAGAUAGCGAUCGCGAGGCUUUCGCUUUUCCUCAAGAUUUUCCGCACACACACACACACAUGUCC- .....((((((((((.(....)))))).....((.((((.......(((((..((...))..))))).......))))...))..................)))))- ( -27.04, z-score = -2.00, R) >dp4.chrXR_group6 10309885 106 + 13314419 UUCAAGGACAGCCCAGCGAAAGUGGGCUCAUUCAUUCUUUCUAGAUAGCGAUCGCGAGGCUUUCGCUUUUCCUCAAGAUUUUCCGCAAACACACACACACAUGUCC- .....((((((((((.(....))))))........((((.......(((((..((...))..))))).......)))).......................)))))- ( -26.84, z-score = -2.26, R) >droEre2.scaffold_4784 22530569 100 - 25762168 UUCGAGGGU-GCCAAGCGAAAAUUGGCC--UUGAUUCUUCCGAGG-AUCGAUGAUAA-GCUUUCACUGAGGC-CGUUGCAAAGUGCAAGUACGCACAAAC-CGUCAA ...((.(((-.....((((....(((((--((((((((....)))-)))..(((...-....)))..)))))-))))))...((((......))))..))-).)).. ( -31.50, z-score = -1.54, R) >droYak2.chr3L 23366049 98 - 24197627 UUCGAGGGU-GGCAAGCAAAAGCCGGCC--UUGAUUCUUUCGAGG-ACCGAUGAUGA-GCUUUCGCUGGGA--CGUUGC-AAGUGCGAGUACUAACAAAC-CGUCAA ...((.(((-....(((.((((((((((--((((.....))))))-.))).......-))))).)))((.(--(.((((-....)))))).)).....))-).)).. ( -29.91, z-score = -0.36, R) >droSec1.super_11 2785097 97 - 2888827 UUCGAGGGU-GCCAAGCGAAAGUCGGCC--UUGAUUCUUGCGAGG-AUCGA---UAA-GCUUUCGCUGGGGC-AGUUGCAAAGUGCGGGUACUCAUAAAC-CGUCGG ..(((.(((-(((.((((((((((((((--(((.......)))))-.))).---...-)))))))))..)))-(.((((.....)))).)........))-).))). ( -36.90, z-score = -2.03, R) >droSim1.chr3L 22155000 76 - 22553184 UUCGAGGGU-GCCAAGCGAAAGUCGGCC--UUGAUUCUUCCGAGG-AUCGA---UAA-GCUUUCGCUGGGGC-CGUCAUAAACGG---------------------- ...((.(((-.(((.(((((((((((((--(((.......)))))-.))).---...-))))))))))).))-).))........---------------------- ( -32.70, z-score = -2.99, R) >consensus UUCGAGGGU_GCCAAGCGAAAGUCGGCC__UUGAUUCUUCCGAGG_AUCGAU__UAA_GCUUUCGCUGGGGC_CGUUACAAAGUGCAAGUACUCACAAAC_CGUCA_ ..............(((((((((.......(((..(((....)))...))).......)))))))))........................................ (-10.55 = -10.80 + 0.25)

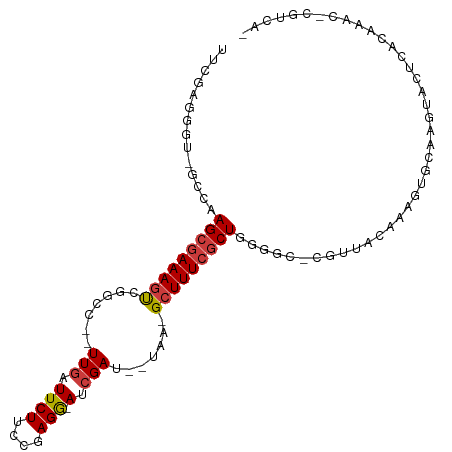
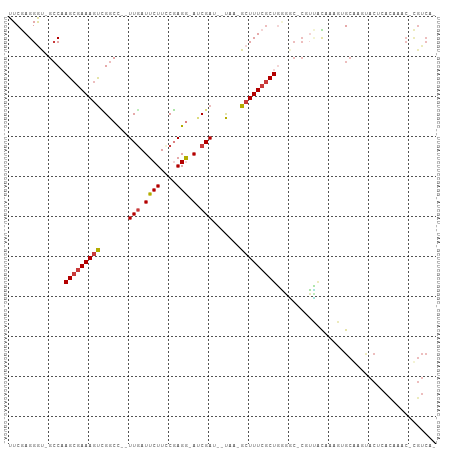
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:56 2011