| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,844,425 – 22,844,501 |
| Length | 76 |
| Max. P | 0.975044 |

| Location | 22,844,425 – 22,844,501 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 72.71 |
| Shannon entropy | 0.50489 |
| G+C content | 0.59111 |
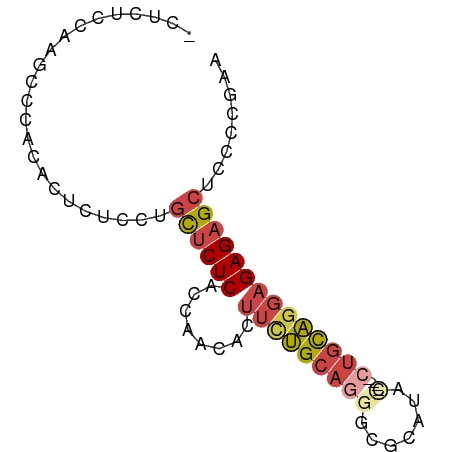
| Mean single sequence MFE | -19.11 |
| Consensus MFE | -12.00 |
| Energy contribution | -11.87 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975044 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
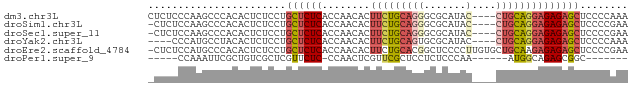
>dm3.chr3L 22844425 76 + 24543557 CUCUCCCAAGCCCACACUCUCCUGCUCUCACCAACACUUCUGCAGGGCGCAUAC----CUGCAGGAGAGAGCUCCCCAAA .......................((((((........(((((((((.......)----))))))))))))))........ ( -22.80, z-score = -2.61, R) >droSim1.chr3L 22117318 75 + 22553184 -CUCUCCAAGCCCACACUCUCCUGCUCUCACCAACACUUCUGCAGGGCGCAUAC----CUGCAGGAGAGAGCUCCCCGAA -......................((((((........(((((((((.......)----))))))))))))))........ ( -22.80, z-score = -2.18, R) >droSec1.super_11 2746981 75 + 2888827 -CUCUCCAAGCCCACACUCUCCUGCUCUCACCAACACUUCUGCAGGGCGCAUAC----CUGCAGGAGAGAGCUCCCCGAA -......................((((((........(((((((((.......)----))))))))))))))........ ( -22.80, z-score = -2.18, R) >droYak2.chr3L 23320471 72 + 24197627 ----CCCAUGCCUACACUCUCCUGCUCUCACCAACACUUCUGCAGUGCGCAUAC----CUGCAGGAGAGAGCUCCCCAAA ----............(((((((((.........((((.....)))).......----..)))))))))........... ( -19.97, z-score = -2.68, R) >droEre2.scaffold_4784 22483788 79 + 25762168 -CUCUCCAUGCCCACACUCUCCUGCUCUCACCAACACUUCUGCACGGCUCCCCUUGUGCUGCAAGAGAGAGCUCCCCGAA -((((((.(((.((((....(((((................))).)).......))))..))).).)))))......... ( -14.79, z-score = -0.36, R) >droPer1.super_9 2288587 61 - 3637205 -----CCAAAUUCGCUGUCGCUCGUUCUC-CCAACUCGUUCGCUCCUCUCCCAA------AUGGCAGAGCGGC------- -----...........((((((((((...-..)))......(((..........------..))).)))))))------- ( -11.50, z-score = -0.41, R) >consensus _CUCUCCAAGCCCACACUCUCCUGCUCUCACCAACACUUCUGCAGGGCGCAUAC____CUGCAGGAGAGAGCUCCCCGAA ................(((((((((...................................)))))))))........... (-12.00 = -11.87 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:53 2011