| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,793,348 – 22,793,465 |
| Length | 117 |
| Max. P | 0.638964 |

| Location | 22,793,348 – 22,793,465 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.39 |
| Shannon entropy | 0.62561 |
| G+C content | 0.65041 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -23.69 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.37 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
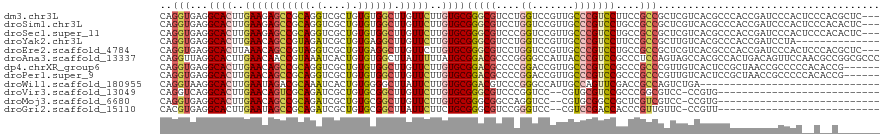
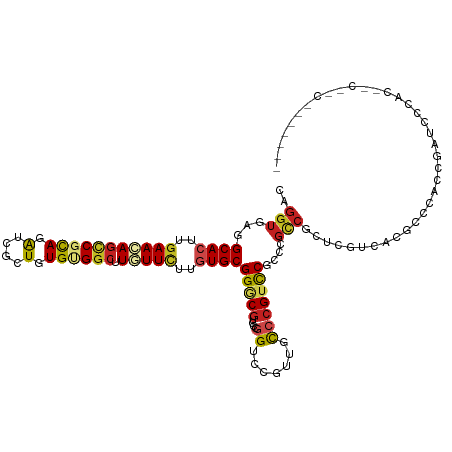
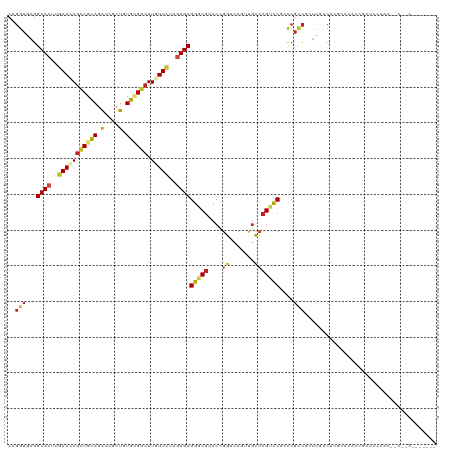
>dm3.chr3L 22793348 117 + 24543557 CAGGUGAGGCACUUGAAGAGCCGCAGGUCGCUGUGUGGCUUGUUCUUGUGCGGGCGUCCUGGUCCGUUGCCCGUCCUUCCGCCGCUCGUCACGCCCACCGAUCCCACUCCCACGCUC--- ..((((.(((....(((((((((((.(....).)))))))).)))..(((((((((.(..((..((.....))..))...).)))))).))))))))))..................--- ( -41.20, z-score = 0.23, R) >droSim1.chr3L 22077941 117 + 22553184 CAGGUGAGGCACUUGAAGAGCCGCAGGUCGCUGUGUGGCUUGUUCUUGUGCGGGCGUCCUGGUCCGUUGCCCGUCCUGCCGCCGCUCGUCACGCCCACCGAUCCCACUCCCACACUC--- ..((.(((((((..(((((((((((.(....).)))))))).)))..))))((((((..((..(.((.((.......)).)).)..))..))))))..........)))))......--- ( -42.80, z-score = 0.03, R) >droSec1.super_11 2709627 117 + 2888827 CAGGUGAGGCACUUGAAGAGCCGCAGGUCGCUGUGUGGCUUGUUCUUGUGCGGGCGUCCCGGUCCGUUGCCCGUCCUGCCGCCGCUCGUCACGCCCACCGAUCCCACUCCCACACUC--- ..((.(((((((..(((((((((((.(....).)))))))).)))..))))((((((..((..(.((.((.......)).)).)..))..))))))..........)))))......--- ( -45.40, z-score = -0.72, R) >droYak2.chr3L 23280500 106 + 24197627 CAGGUGAGGCACUUGAACAGCCGUAGAUCGCUGUGAGGCUUGUUCUUGUGCGGGCGUCCUGGUCCGUUGCCCGUCCUUCCGCCGCUUGUCACGCCCACCGAUCCUA-------------- ..((((.(((....((((((((.(((....)))...))).)))))..(((((((((.(..((..((.....))..))...).)))))).)))))))))).......-------------- ( -37.00, z-score = -0.41, R) >droEre2.scaffold_4784 22447522 117 + 25762168 CAGGUGAGGCACUUAAACAGCCGUAGGUCGCUGUGAGGCUUGUUCUUGUGCGGGCGUCCUGGUCCGUUGCCCGUCCUGCCGCCGCUCGUCACGCCCACCGAUCCCACUCCCACGCUC--- ..((.(((((((...(((((((.(((....)))...))).))))...))))((((((..((..(.((.((.......)).)).)..))..))))))..........)))))......--- ( -35.90, z-score = 1.41, R) >droAna3.scaffold_13337 22549645 120 + 23293914 CAGGUUAGGCACUUGAACAACCGUAAAUCACUGUGUGGCUUAUUUUUAUGCGGACGCCCGGGGCCAUUACCCGUCCGCCCUCCAGUAGCCACGCCACUGACAGUUCCAACGCCGGCGCCC ..(((..(((....((((....((.....)).((((((((.........((((((....(((.......)))))))))........))))))))........))))....)))...))). ( -37.93, z-score = -0.58, R) >dp4.chrXR_group6 10221408 114 - 13314419 CAGGUGAGGCACUUGAACAGCCGCAGGUCGCUGUGUGGCUUGUUCUUGUGCGGACGCCCCGGACCGUUGCCCGUCCGCCCGCCCGUUGUCACUCCGCUAACCGCCCCCACACCG------ ..((((.(((((..(((((((((((.(....).)))))).)))))..))(((((.((..(((((........)))))...))..((....))))))).....)))....)))).------ ( -41.30, z-score = -0.35, R) >droPer1.super_9 2248401 114 - 3637205 CAGGUGAGGCACUUGAACAGCCGCAGGUCGCUGUGUGGCUUGUUCUUGUGCGGACGCCCCGGACCGUUGCCCGUCCGCCCGCCCGUUGUCACUCCGCUAACCGCCCCCACACCG------ ..((((.(((((..(((((((((((.(....).)))))).)))))..))(((((.((..(((((........)))))...))..((....))))))).....)))....)))).------ ( -41.30, z-score = -0.35, R) >droWil1.scaffold_180955 1911197 90 - 2875958 CAGGUAAGGCACUUGAAUAGACGCAAAUCACUGUGGGGCUUAUUCUUGUGCGGACGUCCCGGGCCAUUGCCAGUUCGACCGCCAGUCUGA------------------------------ ..(((...((((..((((((.((((......))))...).)))))..))))((.((..(..(((....))).)..)).))))).......------------------------------ ( -25.90, z-score = 0.60, R) >droVir3.scaffold_13049 10801359 90 - 25233164 CAGGUCAGGCACUUGAACAGUCGCAGAUCGCUGUGCGGCUUGUUCUUGUGCGGGCGUCCCGGUCC--CGUGCGUCCGCCCGGCGUCC-CCGUG--------------------------- ..((....((((..(((((((((((.(....).)))))).)))))..))))(((((((.(((.(.--.....).)))...)))))))-))...--------------------------- ( -38.20, z-score = -1.06, R) >droMoj3.scaffold_6680 12857832 90 + 24764193 CAGGUGAGGCACUUGAACAGCCGCAGAUCGCUGUGCGGCUUGUUCUUGUGCGGGCGGCCAGGUCC--CGUGCGGCCGCUCGUCGUCC-CCGUG--------------------------- ..((.((.((((..(((((((((((.(....).)))))).)))))..))))((((((((.(....--)....))))))))....)).-))...--------------------------- ( -44.90, z-score = -2.00, R) >droGri2.scaffold_15110 15782560 90 + 24565398 CACGUGAGGCACUUGAAUAGCCGCAGAUCGCUGUGCGGCUUAUUCUUCUGCGGGCGUCCGGGUCC--CGUCCGACCACCCGUUGUUC-CCGUU--------------------------- .(((.(..(((...(((((((((((.(....).)))))).)))))....(((((.(..((((...--..))))..).))))))))..-)))).--------------------------- ( -33.80, z-score = -1.30, R) >consensus CAGGUGAGGCACUUGAACAGCCGCAGAUCGCUGUGUGGCUUGUUCUUGUGCGGGCGUCCCGGUCCGUUGCCCGUCCGCCCGCCGCUCGUCACGCCCACCGAUCCCAC__C__C_______ ..(((...((((..(((((((((((.(....).)))))).)))))..))))(((((....((.......)))))))....)))..................................... (-23.69 = -23.93 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:46 2011