
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,753,089 – 22,753,182 |
| Length | 93 |
| Max. P | 0.500000 |

| Location | 22,753,089 – 22,753,182 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.64 |
| Shannon entropy | 0.59596 |
| G+C content | 0.62796 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -14.22 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
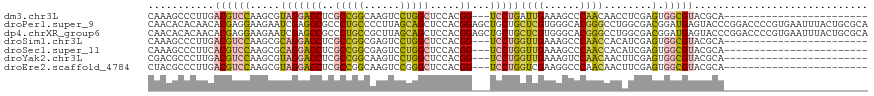
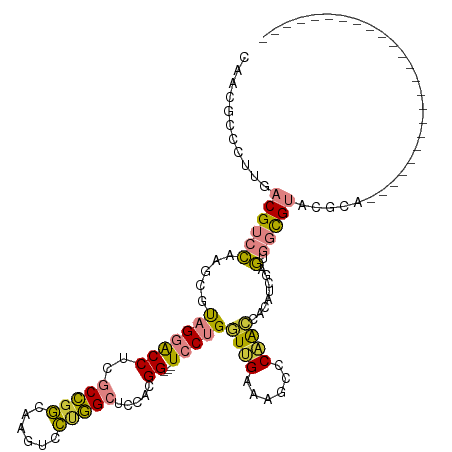
>dm3.chr3L 22753089 93 + 24543557 CAAAGCCCUUGACGUCCAAGCGUAGGACCUCGCCGGCAAGUCCUGGCUCCACGG---UCCUGAUUGAAAGCCCAACAACCUCGAGUGGCGUACGCA------------------------ ....(((((((((((....)))(((((((..(((((......))))).....))---)))))..................))))).))).......------------------------ ( -28.00, z-score = -0.39, R) >droPer1.super_9 2620561 120 - 3637205 CAACACACAACACGAGGAAGAAUCGAGCCGCCCUGCCCCUUAGCAGCUCCACGGAGCUGCUGCUCGUGGGCACGGGCCUGGCGACGGAUGAGUACCCGGACCCCGUGAAUUUACUGCGCA ..........((((.((.........(((((((((((((.(((((((((....)))))))))...).))))).))))..)))..(((........)))..)).))))............. ( -49.90, z-score = -2.74, R) >dp4.chrXR_group6 10594166 120 - 13314419 CAACACACAACACGAGGAAGAAUCGAGCCGCCCUGCCGCUUAGCAGCUCCACGGAGCUGCUGCUCGUGGGCACGGGCCUGGCGACGGAUGAGUACCCGGACCCCGUGAAUUUACUGCGCA ..........((((.((.........(((((((((((((.(((((((((....)))))))))...)).)))).))))..)))..(((........)))..)).))))............. ( -47.20, z-score = -1.61, R) >droSim1.chr3L 22041042 93 + 22553184 CAAAGCCCUUGACGUCCAAGCGCAGGACCUCGCCGGCGAGUCCUGGCUCCACGG---UCCUGGUUGAAAGCCCAACCACAUCGAGUGGCGUACGCA------------------------ ....(((((((((((...(((.((((((.(((....))))))))))))..))).---...((((((......))))))..))))).))).......------------------------ ( -37.50, z-score = -2.13, R) >droSec1.super_11 2673290 93 + 2888827 CAAAGCCCUUCACGUCCAAGCGCAGGACCUCGCCGGCGAGUCCUGGCUCCACGG---UCCUGGUUGAAAGCCCAACCACAUCGAGUGGCGUACGCA------------------------ ....(((.(((.(((...(((.((((((.(((....))))))))))))..))).---...((((((......))))))....))).))).......------------------------ ( -34.40, z-score = -1.53, R) >droYak2.chr3L 23244781 93 + 24197627 CGACGCCCUUGACGUCCAAGCGUAGGACCUCGCCGGCAAGUCCUGGCUCCACGG---UCCUGGUUGAAAGUCCAACAACUUCGAGUGGCGUACGCA------------------------ ((((((((((((.((.......(((((((..(((((......))))).....))---)))))((((......)))).)).))))).))))).))..------------------------ ( -35.40, z-score = -1.59, R) >droEre2.scaffold_4784 22405034 93 + 25762168 CUACGCCCUUGACGUCCAAGCGUAGGACCUCGCCGGCAAGUCCGGGCUCCACGG---UCCUGGUCGAAGGCCCAACAACUUCGAGUGGCGUACGCA------------------------ .(((((((((((.((((.......))))......(((..(.(((((((....))---.))))).)....)))........))))).))))))....------------------------ ( -33.60, z-score = -0.22, R) >consensus CAACGCCCUUGACGUCCAAGCGUAGGACCUCGCCGGCAAGUCCUGGCUCCACGG___UCCUGGUUGAAAGCCCAACCACAUCGAGUGGCGUACGCA________________________ ...................(((((.(..((((((((......)))))...............((((......))))......)))...).)))))......................... (-14.22 = -13.60 + -0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:43 2011