
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,171,096 – 6,171,204 |
| Length | 108 |
| Max. P | 0.561471 |

| Location | 6,171,096 – 6,171,204 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.54372 |
| G+C content | 0.39638 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -10.17 |
| Energy contribution | -10.34 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6171096 108 - 23011544 --UCGUCUUUCUUGCGAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGAGGUUUCAGCCCAGG----ACUCCGUGGCCCUGAAUG --(((((...((((((((((((.((((......))))))))))))))))..))).......(((((.....)))))))...(((((.(((.(----....).)))..))))).. ( -31.60, z-score = -2.50, R) >droSim1.chr2L 5963886 108 - 22036055 --UCGUCUUUCUUGCGAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGAGGUUUCAGCCCAGG----ACUCCGUGGCACAGAAUG --..(((...((((((((((((.((((......))))))))))))))))..)))..............(((((.(((..((((.......))----))..))).)))))..... ( -33.20, z-score = -2.99, R) >droSec1.super_5 4242358 108 - 5866729 --UCGUCUUUCUUGCGAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGAGGUUUCAGCCCAGG----ACUCCGAGGCACAGAAUG --..(((...((((((((((((.((((......))))))))))))))))..)))..............(((((..((..((((.......))----))..))..)))))..... ( -32.10, z-score = -2.67, R) >droYak2.chr2L 15584531 108 + 22324452 --UCGUCUUUCUUGCGAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGAGGUUUCAGCCCAGG----ACUCCGUGGCGCAGAAUG --..(((...((((((((((((.((((......))))))))))))))))..)))((((...(((((.....))))).........(((((.(----....).))).)).)))). ( -33.20, z-score = -2.73, R) >droEre2.scaffold_4929 15098193 112 - 26641161 --UCGUCUUUCUUGCGAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGAGGUUUCAGCCCAGGACACACUCCGUGGCGCAGAGUG --..(((...((((((((((((.((((......))))))))))))))))..)))((((...(((((.....))))).........((((((((.....))).))).)).)))). ( -34.90, z-score = -2.69, R) >droAna3.scaffold_12916 8148988 106 + 16180835 --UUUUCUUUCUAGCACUUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAAUUGUUAUGUGAAACGUGAAA-UUG-AGACUGGAGGUCCUGGCGCUGUG---- --...........(((..((((.((((......))))))))..)))..((..(...((((.....(((((.....)))))..-)))-)(((.....))))..))......---- ( -16.50, z-score = 1.55, R) >droWil1.scaffold_180708 5000437 87 + 12563649 UUGCUGCUCCCCAACAAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGUGGUAUAGAG--------------------------- .(((..(.......((((((((.((((......))))))))))))...(((((((((((....)).))))).)))))..))).....--------------------------- ( -19.20, z-score = -1.07, R) >droVir3.scaffold_12963 17131455 91 + 20206255 --UUCUUAUCCCAGUAAUUCAAAUUAAUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUCUAAUGUGCAACGUGAACUUCGUGGACUG--------------------- --......(((((((((((......)))))))...((((((((((((.(((((.((.....)).))))).))))).)))))))..).)))...--------------------- ( -18.10, z-score = -1.64, R) >droGri2.scaffold_15252 14513397 90 + 17193109 --UUCUUGUGCUGGUAAUUCAAAUUAUUUAUUUAUGGUUCAUUUGCGAGUUGGCAUUCAAAGCUGUAAUGUGCAGUGUGAAC-UCGUGGACUG--------------------- --....((.((((((((..........)))))...))).))((..((((((.(((((....((......))..))))).)))-)))..))...--------------------- ( -18.70, z-score = -0.14, R) >consensus __UCGUCUUUCUUGCGAGUGAAAUUAUUUAUUUAUAGUUCAUUUGCGAGUUGGCAUUCAAAGUUGUAAUGUGCAACGAGGUUUCAGCCCAGG____ACUCCGUGGC_CAGA_UG .............((((.((((.((((......)))))))).))))..(((((((((((....)).))))).))))...................................... (-10.17 = -10.34 + 0.18)

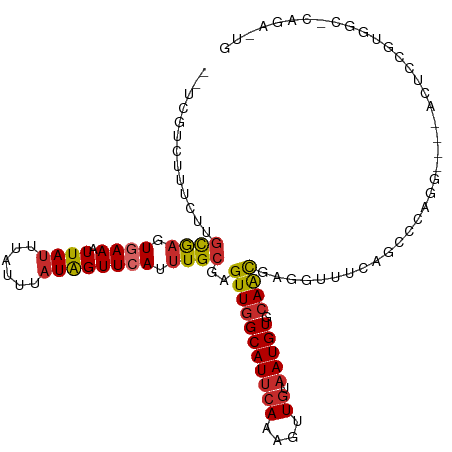
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:34 2011