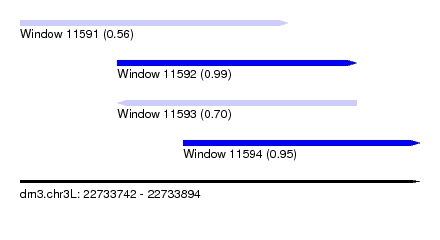
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,733,742 – 22,733,894 |
| Length | 152 |
| Max. P | 0.992165 |

| Location | 22,733,742 – 22,733,844 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 60.54 |
| Shannon entropy | 0.89220 |
| G+C content | 0.61078 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.71 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
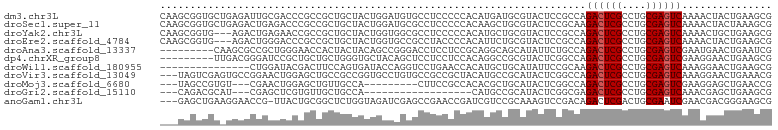
>dm3.chr3L 22733742 102 + 24543557 CAAGCGGUGCUGAGAUUGCGACCCGCCGCUGCUACUGGAUGUGCCUCCCCCACAUGAUGCGUACUCCGCCAGACUCGCCUGCGAGUCAAAACUACUGAAGCG ..(((((((....(....)....)))))))(((...(((.(((((((........)).).)))))))....((((((....))))))...........))). ( -30.20, z-score = -1.30, R) >droSec1.super_11 2654537 102 + 2888827 CAAGCGGUGCUGAGACUGAGACCCGCCGCUGCUACUGGAUGCGCCUCCCCCACAAGCUGCGUACUCCGCAAGACUCGCCUGCGAGUCAAAACUACUAAAGCG ..(((((((..............)))))))(((...((((((((..(........)..))))).)))....((((((....))))))...........))). ( -30.94, z-score = -1.27, R) >droYak2.chr3L 23224757 99 + 24197627 CAAGCGGUG---AGACUGAGAACCGCCGCUGCUACUGGUGGCGCCUCCCCCACAUGCUGCGUACUCCGCCAGACUCGCCUGCGAGUCAAAACUGCUGAAGCG ..(((((((---.(........))))))))(((..(((((((((..(........)..)))....))))))((((((....))))))...........))). ( -33.10, z-score = -0.88, R) >droEre2.scaffold_4784 22385098 99 + 25762168 CAAGCGGUG---AGACUGGGACCCGCCGCUGCUACUGGUGCCGCCUACCCCACAUUCUGCGUACUCCGCCAGACUCGCCUGCGAGUCAAAACUACUGAAGCG ..(((((((---.(........))))))))(((..(((((.(((..............))).....)))))((((((....))))))...........))). ( -29.74, z-score = -0.48, R) >droAna3.scaffold_13337 22504101 93 + 23293914 ---------CAAGCGCCGCUGGGAACCACUACUACAGCCGGGACCUCCUCCGCAGGCAGCAUAUUCUGCCAGACUCGCCUGCGAGUCGAAUGAACUGAAUCG ---------...(((..((((.............)))).(((....))).))).(((((......))))).((((((....))))))............... ( -28.02, z-score = -0.74, R) >dp4.chrXR_group8 413000 93 + 9212921 ---------UUGACGGGAUCCGCUGCUGCUGGGUGCUACAGCUCCUCCUCCACAGGCCGCGUACUCGGCCAGACUCGCCUGCGAGUCGAAGGAACUGAAGCG ---------........((((((....)).))))(((.(((.((((.(......(((((......))))).((((((....))))))).)))).))).))). ( -37.60, z-score = -1.11, R) >droWil1.scaffold_180955 496174 87 - 2875958 ---------------CUGGAUACGACUUCCAGUGAUACCAGGUCCUGAACCACAUGCUGCAUAUUCCGCAAGACUCGCCUGCGAGUCAAAGGAACUGAAGCG ---------------(((((.......)))))......(((.((((...........(((.......))).((((((....))))))..)))).)))..... ( -25.60, z-score = -1.99, R) >droVir3.scaffold_13049 10702916 99 - 25233164 ---UAGUCGAGUGCCGGAACUGGAGCUGCCGCCGGUGCCUGUGCCGCCGCUACAUGCCGCAUACUCGGCCAGACUCGCCUGCGAGUCAAAGGAACUGAAACG ---..((((((((.(((...((.(((.((.(((((...))).)).)).))).))..)))..))))))))..((((((....))))))............... ( -35.90, z-score = -0.60, R) >droMoj3.scaffold_6680 12769577 87 + 24764193 ---UAGCCGUGU---CGAACUGGAGCUGUUGCCA---------CUUCCGCCACACGCUGCAUACUCGGCCAGACUCGCCUGCGAGUCGAAGGAGCUGAACCG ---((((((.((---(((..((.((((((.((..---------.....)).))).))).))...)))))).((((((....))))))....).))))..... ( -28.70, z-score = -0.46, R) >droGri2.scaffold_15110 15703002 78 + 24565398 ---CAGACGCAU---CGAGCUCGUGUUGCUGCCA------------------CAUGCCGCAUACUCGGCGAGACUCGCCUGCGAGUCAAACGAGCUGAAGCG ---....(((.(---(.(((((((..........------------------..(((((......))))).((((((....))))))..))))))))).))) ( -32.80, z-score = -2.25, R) >anoGam1.chr3L 37149896 98 + 41284009 ---GAGCUGAAGGAACCG-UUACUGCGGCUCUGGUAGAUCGAGCCGAACCGAUCGUCCGCAAAGUCCGACAGACUCGACUGCGAAUCGAACGACGGGAAGCG ---((((((..(((.(((-(....))))....((..(((((........)))))..))......)))..))).)))....((...(((.....)))...)). ( -28.40, z-score = 0.71, R) >consensus ___GAGGUGC_GAGACUGACUCCCGCCGCUGCUACUGGCGGCGCCUCCCCCACAUGCUGCAUACUCCGCCAGACUCGCCUGCGAGUCAAAAGAACUGAAGCG .......................................................................((((((....))))))............... ( -9.62 = -9.71 + 0.09)

| Location | 22,733,779 – 22,733,870 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.59657 |
| G+C content | 0.59104 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
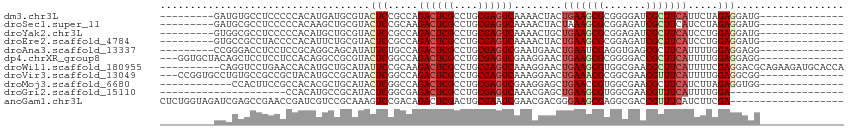
>dm3.chr3L 22733779 91 + 24543557 ---------GAUGUGCCUCCCCCACAUGAUGCGUACUCCGCCAGACUCGCCUGCGAGUCAAAACUACUGAAGCGCGGGGAUCGCUUCAUUCUAGAGGAUG-------------- ---------......((((......((((.(((..(((((((.((((((....))))))....(....)..).))))))..))).))))....))))...-------------- ( -31.40, z-score = -2.29, R) >droSec1.super_11 2654574 91 + 2888827 ---------GAUGCGCCUCCCCCACAAGCUGCGUACUCCGCAAGACUCGCCUGCGAGUCAAAACUACUAAAGCGCGGAGAUCGCUUCAUCCUAGAGGAUG-------------- ---------(((((((..(........)..)))).((((((..((((((....))))))....((.....)).)))))))))....(((((....)))))-------------- ( -29.90, z-score = -2.62, R) >droYak2.chr3L 23224791 91 + 24197627 ---------GUGGCGCCUCCCCCACAUGCUGCGUACUCCGCCAGACUCGCCUGCGAGUCAAAACUGCUGAAGCGCGGAGAUCGCUUCAUCCUGGAGGAUG-------------- ---------......(((((.....(((..(((..((((((..((((((....))))))......((....))))))))..)))..)))...)))))...-------------- ( -37.10, z-score = -2.79, R) >droEre2.scaffold_4784 22385132 91 + 25762168 ---------GUGCCGCCUACCCCACAUUCUGCGUACUCCGCCAGACUCGCCUGCGAGUCAAAACUACUGAAGCGCGGAGAUCGCUUCAUCCUGGAGGAUG-------------- ---------..........(((((......(((..(((((((.((((((....))))))....(....)..).))))))..))).......))).))...-------------- ( -29.42, z-score = -1.98, R) >droAna3.scaffold_13337 22504129 91 + 23293914 ---------CCGGGACCUCCUCCGCAGGCAGCAUAUUCUGCCAGACUCGCCUGCGAGUCGAAUGAACUGAAUCGAGGUGAGCGCUUCAUUUUGGAGGAGG-------------- ---------......((((((((...(((((......))))).(.(((((((.(((.(((.......))).)))))))))))..........))))))))-------------- ( -39.80, z-score = -2.96, R) >dp4.chrXR_group8 413022 97 + 9212921 ---GGUGCUACAGCUCCUCCUCCACAGGCCGCGUACUCGGCCAGACUCGCCUGCGAGUCGAAGGAACUGAAGCGCGGGGACCGCUUCAUUUUGGAGGAGG-------------- ---...((....)).((((((((...(((((......))))).((((((....))))))....(((.(((((((.(....)))))))).)))))))))))-------------- ( -49.70, z-score = -3.76, R) >droWil1.scaffold_180955 496197 104 - 2875958 ----------CAGGUCCUGAACCACAUGCUGCAUAUUCCGCAAGACUCGCCUGCGAGUCAAAGGAACUGAAGCGUGGCGAACGCUUCAUUUUCGAGGACGCAGAAGAUGCACCA ----------..(((.....)))...((.(((((.(((.((..((((((....))))))...((((.((((((((.....)))))))).))))......)).))).))))).)) ( -38.90, z-score = -3.03, R) >droVir3.scaffold_13049 10702944 97 - 25233164 ---CCGGUGCCUGUGCCGCCGCUACAUGCCGCAUACUCGGCCAGACUCGCCUGCGAGUCAAAGGAACUGAAACGCGGCGAACGUUUCAUUUUGGAGGCGG-------------- ---..((((....))))(((((.....((((......))))..((((((....))))))..............)))))...((((((......)))))).-------------- ( -36.20, z-score = -0.73, R) >droMoj3.scaffold_6680 12769602 88 + 24764193 ------------CCACUUCCGCCACACGCUGCAUACUCGGCCAGACUCGCCUGCGAGUCGAAGGAGCUGAACCGUGGCGAACGCUUCAUCUUAGAGGUGG-------------- ------------.......((((((..((((......))))..((((((....))))))..............))))))..((((((......)))))).-------------- ( -32.40, z-score = -1.59, R) >droGri2.scaffold_15110 15703027 74 + 24565398 ---------------------CCACAUGCCGCAUACUCGGCGAGACUCGCCUGCGAGUCAAACGAGCUGAAGCGUGGCGAACGUUUCAUUUUGGA------------------- ---------------------.....(((((......))))).((((((....))))))...((((.((((((((.....)))))))).))))..------------------- ( -28.70, z-score = -2.37, R) >anoGam1.chr3L 37149920 95 + 41284009 CUCUGGUAGAUCGAGCCGAACCGAUCGUCCGCAAAGUCCGACAGACUCGACUGCGAAUCGAACGACGGGAAGCGAGGCGACCGUUUCAUCUUCGA------------------- ....((..(((((........)))))..))(((.((((.....))))....)))...(((((.((...((((((.(....))))))).)))))))------------------- ( -26.10, z-score = 1.60, R) >consensus _________GCGGCGCCUCCCCCACAUGCUGCAUACUCCGCCAGACUCGCCUGCGAGUCAAAAGAACUGAAGCGCGGCGAACGCUUCAUUUUGGAGGAGG______________ ...................................(((.....((((((....))))))........(((((((.......))))))).....))).................. (-17.48 = -17.79 + 0.31)
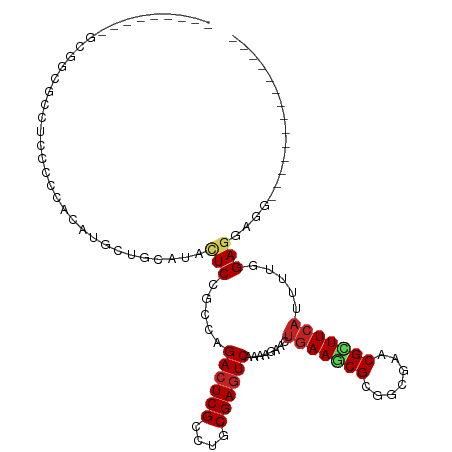
| Location | 22,733,779 – 22,733,870 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.59657 |
| G+C content | 0.59104 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -13.61 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22733779 91 - 24543557 --------------CAUCCUCUAGAAUGAAGCGAUCCCCGCGCUUCAGUAGUUUUGACUCGCAGGCGAGUCUGGCGGAGUACGCAUCAUGUGGGGGAGGCACAUC--------- --------------..(((((((..((((.(((..(.(((((((.....)))...((((((....))))))..)))).)..))).)))).)))))))........--------- ( -34.60, z-score = -1.80, R) >droSec1.super_11 2654574 91 - 2888827 --------------CAUCCUCUAGGAUGAAGCGAUCUCCGCGCUUUAGUAGUUUUGACUCGCAGGCGAGUCUUGCGGAGUACGCAGCUUGUGGGGGAGGCGCAUC--------- --------------(((((....)))))..(((.(((((.(((...(((......((((((....)))))).((((.....))))))).))))))))..)))...--------- ( -38.50, z-score = -2.46, R) >droYak2.chr3L 23224791 91 - 24197627 --------------CAUCCUCCAGGAUGAAGCGAUCUCCGCGCUUCAGCAGUUUUGACUCGCAGGCGAGUCUGGCGGAGUACGCAGCAUGUGGGGGAGGCGCCAC--------- --------------..(((((((.(.((..(((..((((((((....))......((((((....))))))..))))))..)))..))).)))))))........--------- ( -41.00, z-score = -2.38, R) >droEre2.scaffold_4784 22385132 91 - 25762168 --------------CAUCCUCCAGGAUGAAGCGAUCUCCGCGCUUCAGUAGUUUUGACUCGCAGGCGAGUCUGGCGGAGUACGCAGAAUGUGGGGUAGGCGGCAC--------- --------------(((((....)))))..((...(((((((.(((.........((((((....))))))..(((.....))).))))))))))...)).....--------- ( -34.30, z-score = -1.22, R) >droAna3.scaffold_13337 22504129 91 - 23293914 --------------CCUCCUCCAAAAUGAAGCGCUCACCUCGAUUCAGUUCAUUCGACUCGCAGGCGAGUCUGGCAGAAUAUGCUGCCUGCGGAGGAGGUCCCGG--------- --------------((((((((.......((.((((.((((((.((.........)).))).))).))))))(((((......)))))...))))))))......--------- ( -37.20, z-score = -2.71, R) >dp4.chrXR_group8 413022 97 - 9212921 --------------CCUCCUCCAAAAUGAAGCGGUCCCCGCGCUUCAGUUCCUUCGACUCGCAGGCGAGUCUGGCCGAGUACGCGGCCUGUGGAGGAGGAGCUGUAGCACC--- --------------(((((((((...(((((((.......)))))))........((((((....)))))).(((((......)))))..))))))))).((....))...--- ( -47.70, z-score = -3.56, R) >droWil1.scaffold_180955 496197 104 + 2875958 UGGUGCAUCUUCUGCGUCCUCGAAAAUGAAGCGUUCGCCACGCUUCAGUUCCUUUGACUCGCAGGCGAGUCUUGCGGAAUAUGCAGCAUGUGGUUCAGGACCUG---------- ((.(((((.(((((((.....(((..((((((((.....)))))))).)))....((((((....)))))).))))))).))))).))...((((...))))..---------- ( -40.00, z-score = -2.50, R) >droVir3.scaffold_13049 10702944 97 + 25233164 --------------CCGCCUCCAAAAUGAAACGUUCGCCGCGUUUCAGUUCCUUUGACUCGCAGGCGAGUCUGGCCGAGUAUGCGGCAUGUAGCGGCGGCACAGGCACCGG--- --------------(((.................((((((((..((((.....))))..))).)))))(((((((((....(((........))).)))).)))))..)))--- ( -36.30, z-score = -0.67, R) >droMoj3.scaffold_6680 12769602 88 - 24764193 --------------CCACCUCUAAGAUGAAGCGUUCGCCACGGUUCAGCUCCUUCGACUCGCAGGCGAGUCUGGCCGAGUAUGCAGCGUGUGGCGGAAGUGG------------ --------------((((.((......))....((((((((((((((((((..((((((((....)))))).))..)))).)).))).))))))))).))))------------ ( -33.90, z-score = -0.99, R) >droGri2.scaffold_15110 15703027 74 - 24565398 -------------------UCCAAAAUGAAACGUUCGCCACGCUUCAGCUCGUUUGACUCGCAGGCGAGUCUCGCCGAGUAUGCGGCAUGUGG--------------------- -------------------..................(((((((.(((((((...((((((....))))))....))))).)).)))..))))--------------------- ( -24.00, z-score = -0.81, R) >anoGam1.chr3L 37149920 95 - 41284009 -------------------UCGAAGAUGAAACGGUCGCCUCGCUUCCCGUCGUUCGAUUCGCAGUCGAGUCUGUCGGACUUUGCGGACGAUCGGUUCGGCUCGAUCUACCAGAG -------------------(((((((((....((..((...))..)))))).))))).(((.((((((..(.((((..(.....)..)))).)..))))))))).......... ( -28.90, z-score = 1.38, R) >consensus ______________CAUCCUCCAAAAUGAAGCGAUCGCCGCGCUUCAGUUCCUUUGACUCGCAGGCGAGUCUGGCGGAGUAUGCAGCAUGUGGGGGAGGCGCAGC_________ ....................................(((((..............((((((....))))))..((.......)).....))))).................... (-13.61 = -14.05 + 0.44)


| Location | 22,733,804 – 22,733,894 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Shannon entropy | 0.54756 |
| G+C content | 0.57017 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.31 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.949940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22733804 90 + 24543557 ACUCCGCCAGACUCGCCUGCGAGUCAAAACUACUGAAGCGCGGGGAUCGCUUCAUUCUAGAGGAUGCACUGCUGCCGUUCACAUCAGGCG ....((((.((((((....))))))........(((((((((((.(((.(((.......)))))).).)))).))..)))).....)))) ( -29.20, z-score = -0.86, R) >droSec1.super_11 2654599 90 + 2888827 ACUCCGCAAGACUCGCCUGCGAGUCAAAACUACUAAAGCGCGGAGAUCGCUUCAUCCUAGAGGAUGCACUGCUGCUGUUAACAUCAGGCG .((((((..((((((....))))))....((.....)).))))))..(((..(((((....))))).....(((.((....)).)))))) ( -31.70, z-score = -2.63, R) >droYak2.chr3L 23224816 90 + 24197627 ACUCCGCCAGACUCGCCUGCGAGUCAAAACUGCUGAAGCGCGGAGAUCGCUUCAUCCUGGAGGAUGCACUGCUGCCAUUCACAUCAGGCG .((((((..((((((....))))))......((....))))))))...((..(((((....)))))....)).(((..........))). ( -34.10, z-score = -1.98, R) >droEre2.scaffold_4784 22385157 90 + 25762168 ACUCCGCCAGACUCGCCUGCGAGUCAAAACUACUGAAGCGCGGAGAUCGCUUCAUCCUGGAGGAUGCACUGCUGCCGUUCACAUCAGACG .(((((((.((((((....))))))....(....)..).))))))...((..(((((....)))))....))...((((.......)))) ( -28.30, z-score = -1.03, R) >droAna3.scaffold_13337 22504154 90 + 23293914 AUUCUGCCAGACUCGCCUGCGAGUCGAAUGAACUGAAUCGAGGUGAGCGCUUCAUUUUGGAGGAGGCGCUGGAACCGUUCGCAUCUGAUG ....(((..((((((....))))))((((((......))..(((.((((((((.........))))))))...))))))))))....... ( -29.80, z-score = -0.56, R) >dp4.chrXR_group8 413053 90 + 9212921 ACUCGGCCAGACUCGCCUGCGAGUCGAAGGAACUGAAGCGCGGGGACCGCUUCAUUUUGGAGGAGGCACUGCUCCCGUUCGCGUCAGGUG .....(((.((((((....))))))((.((((((((((((.(....))))))))....((((.((...)).)))).)))).).)).))). ( -37.90, z-score = -1.03, R) >droVir3.scaffold_13049 10702975 84 - 25233164 ACUCGGCCAGACUCGCCUGCGAGUCAAAGGAACUGAAACGCGGCGAACGUUUCAUUUUGGAGGCGGCGCUG---CCAUUCAAAUCCG--- .........((((((....))))))...(((..((((..((((((..((((((......)))))).)))))---)..))))..))).--- ( -33.20, z-score = -2.48, R) >droMoj3.scaffold_6680 12769624 84 + 24764193 ACUCGGCCAGACUCGCCUGCGAGUCGAAGGAGCUGAACCGUGGCGAACGCUUCAUCUUAGAGGUGGCGCUG---CCAUUCAAAUCCG--- .........((((((....))))))...(((..((((..(..(((..((((((......)))))).)))..---)..))))..))).--- ( -33.40, z-score = -2.00, R) >droGri2.scaffold_15110 15703040 75 + 24565398 ACUCGGCGAGACUCGCCUGCGAGUCAAACGAGCUGAAGCGUGGCGAACGUUUCAUUUUGGAG------CUG---CCAUUUAAAU------ ....((((.((((((....))))))...((((.((((((((.....)))))))).))))...------.))---))........------ ( -28.40, z-score = -2.40, R) >anoGam1.chr3L 37149954 84 + 41284009 AGUCCGACAGACUCGACUGCGAAUCGAACGACGGGAAGCGAGGCGACCGUUUCAUCUUCGACUCACUGCUGAGCGUGUUUACUG------ (((..((((..((((.(.(.((.(((((.((...((((((.(....))))))).))))))).)).).).))))..)))).))).------ ( -23.80, z-score = 0.44, R) >consensus ACUCCGCCAGACUCGCCUGCGAGUCAAAAGAACUGAAGCGCGGCGAUCGCUUCAUCUUGGAGGAGGCACUGCUGCCGUUCACAUCAG__G .(((.....((((((....))))))........(((((((.......))))))).....)))............................ (-18.29 = -18.31 + 0.02)
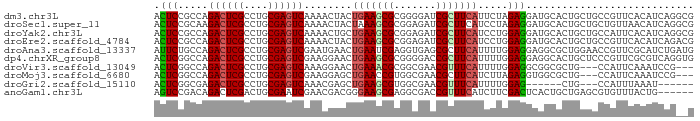
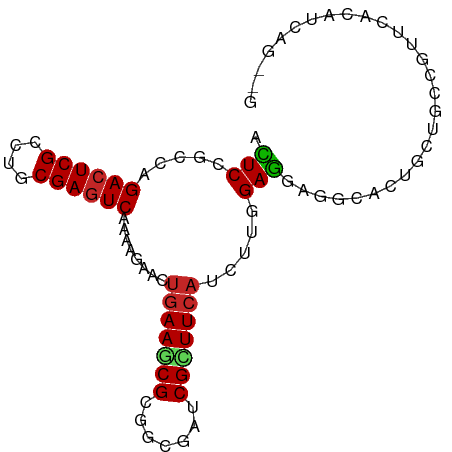
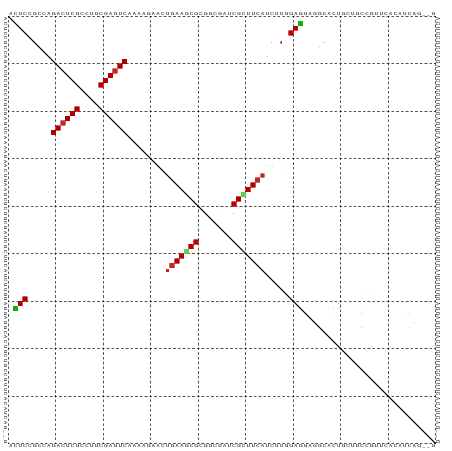
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:40 2011