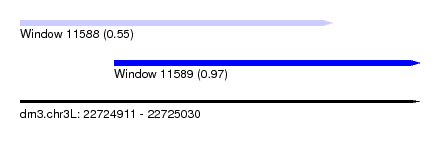
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,724,911 – 22,725,030 |
| Length | 119 |
| Max. P | 0.966053 |

| Location | 22,724,911 – 22,725,004 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 68.05 |
| Shannon entropy | 0.60173 |
| G+C content | 0.41019 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -10.60 |
| Energy contribution | -11.33 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
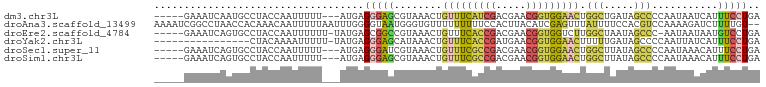
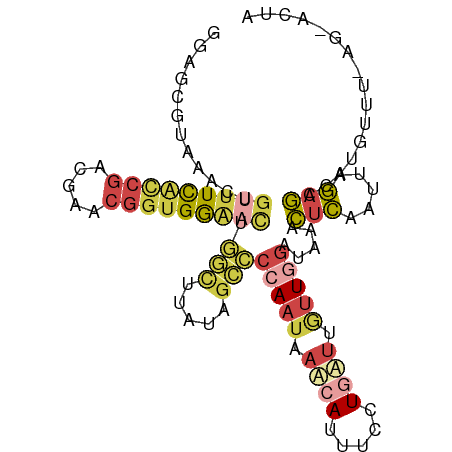
>dm3.chr3L 22724911 93 + 24543557 -----GAAAUCAAUGCCUACCAAUUUUU---AUGAGGGAGCGUAAACUGUUUCAUCGACGAACGGUGGAACUGGCUGAUAGCCCCAAUAAUCAUUUCCUGA -----.......................---....(((.((.((..(.(((((((((.....))))))))).)..))...)))))................ ( -19.60, z-score = -0.26, R) >droAna3.scaffold_13499 254431 99 - 1739075 AAAAUCGGCCUAACCACAAACAAUUUUUAAUUUGGGGUAAUGGGUGUUUUUUUUUCCACUUACAUCGAGUUUAUUUUCCACGUCCAAAAGAUCUUUUGU-- ......((.....))(((((..((((((((((((..((((((((..........)))).))))..))))))...............))))))..)))))-- ( -12.56, z-score = 1.30, R) >droEre2.scaffold_4784 22375410 94 + 25762168 -----GAAAUCAGUGCCUACCAAUUUUUU-UAUGAGCGGCCGUAAACUGUUUCACCGACGAACGGUGGUCUUGGCUAAUAGCCC-AAUAAUAAUGUCCUGA -----....((((..(.............-...((((.(((((....((((.....)))).))))).).)))(((.....))).-.........)..)))) ( -17.60, z-score = 0.49, R) >droYak2.chr3L 23212181 84 + 24197627 ----------------CUACAAAAUUUUU-UAUGAGGGAGCAUAAACUGUUUCACCGAUGAACGGUGGAACUUUUUGAUAGCCCCAAUUAUCAUUUCCUGA ----------------.............-.....(((.((.((((..(((((((((.....)))))))))..))))...)))))................ ( -21.30, z-score = -2.77, R) >droSec1.super_11 2645793 93 + 2888827 -----GAAAUCAGUGCCUACCAAUUUUU---AUGAGGGAUCGUAAACUGUUUCGCCGACGAACGGUGGAACUGGCUUAUAGCCCCAAUAAACAUUUCCUGA -----....((((............(((---((((....)))))))..(((((((((.....))))))))).(((.....)))..............)))) ( -22.10, z-score = -1.11, R) >droSim1.chr3L 22012052 93 + 22553184 -----GAAAUCAGUGCCUACCAAUUUUU---AUGAGGGAGCGUAAACUGUUUCGCCGACGAACGGUGGAACUGGCUUAUAGCCCCAAUAAACAUUUCCUGA -----....((((.(......(((.(((---((..(((.((((((.(((((((((((.....))))))))).)).)))).))))).))))).))).))))) ( -23.50, z-score = -1.33, R) >consensus _____GAAAUCAGUGCCUACCAAUUUUU___AUGAGGGAGCGUAAACUGUUUCACCGACGAACGGUGGAACUGGCUUAUAGCCCCAAUAAACAUUUCCUGA ...................................(((((........(((((((((.....))))))))).(((.....)))...........))))).. (-10.60 = -11.33 + 0.73)
| Location | 22,724,939 – 22,725,030 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 65.05 |
| Shannon entropy | 0.67194 |
| G+C content | 0.40945 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -10.16 |
| Energy contribution | -11.67 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
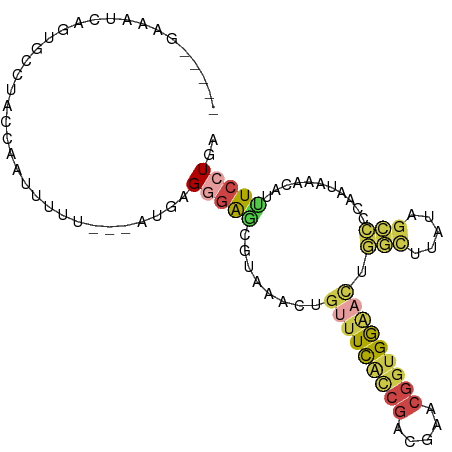
>dm3.chr3L 22724939 91 + 24543557 GGAGCGUAAACUGUUUCAUCGACGAACGGUGGAACUGGCUGAUAGCCCCAAUAAUCAUUUCCUGAUUGUUGGAUAAACUCAAUUAGAGUCA------------- .(((..((....(((((((((.....))))))))).(((.....)))((((((((((.....)))))))))).))..)))...........------------- ( -27.40, z-score = -2.59, R) >droAna3.scaffold_13499 254467 91 - 1739075 GUAAUGGGUGUUUUUUUUUCCACUUACAUCGAGUUUAUUUUCCACGUCCAA--AAGAUCUUUUGUCGGGUUGAGCGAUGAAUCUAUAAUUUUA----------- ((((((((..........)))).)))).(((.(((((....((.((..(((--((....))))).)))).)))))..))).............----------- ( -14.20, z-score = 0.30, R) >droEre2.scaffold_4784 22375440 99 + 25762168 CGGCCGUAAACUGUUUCACCGACGAACGGUGGUCUUGGCUAAUAGCCC-AAUAAUAAUGUCCUGAUAAUUU--CAGGCUCAAU-AGAGCC-UAAAUUAGUACUA .((.(((........((((((.....))))))....(((.....))).-.......))).))...((((((--.((((((...-.)))))-)))))))...... ( -26.40, z-score = -2.01, R) >droYak2.chr3L 23212200 103 + 24197627 GGAGCAUAAACUGUUUCACCGAUGAACGGUGGAACUUUUUGAUAGCCCCAAUUAUCAUUUCCUGAUUGUUGGAUAAACUCAAUUAGAGCC-CGUUUUAGUAAUA ((.((.((((..(((((((((.....)))))))))..))))......(((((.((((.....)))).)))))...............)))-)............ ( -26.80, z-score = -2.69, R) >droSec1.super_11 2645821 104 + 2888827 GGAUCGUAAACUGUUUCGCCGACGAACGGUGGAACUGGCUUAUAGCCCCAAUAAACAUUUCCUGAUUGUUGGAUAAACUCAAUUAGAGCCAUGUUUCAGCACUA ((..........(((((((((.....))))))))).(((.....)))))...((((((..((((((((..(......).))))))).)..))))))........ ( -26.50, z-score = -1.51, R) >droSim1.chr3L 22012080 104 + 22553184 GGAGCGUAAACUGUUUCGCCGACGAACGGUGGAACUGGCUUAUAGCCCCAAUAAACAUUUCCUGAUUGUUGGAUAAACUCACUUAGAGCCAUGUUUCAGCACUA (((((((.....(((((((((.....))))))))).(((((......(((((((.((.....)).))))))).(((......)))))))))))))))....... ( -30.80, z-score = -2.54, R) >consensus GGAGCGUAAACUGUUUCACCGACGAACGGUGGAACUGGCUUAUAGCCCCAAUAAACAUUUCCUGAUUGUUGGAUAAACUCAAUUAGAGCCAUGUUU_AG_ACUA ............(((((((((.....))))))))).(((.....)))(((((.((((.....)))).))))).....(((.....)))................ (-10.16 = -11.67 + 1.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:36 2011