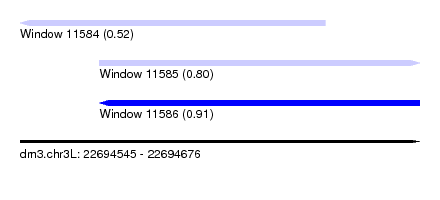
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,694,545 – 22,694,676 |
| Length | 131 |
| Max. P | 0.905815 |

| Location | 22,694,545 – 22,694,645 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Shannon entropy | 0.43427 |
| G+C content | 0.59463 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
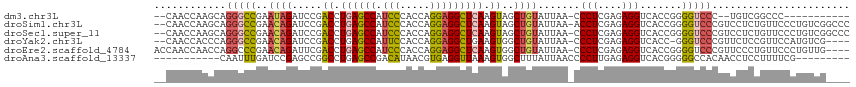
>dm3.chr3L 22694545 100 - 24543557 --CAACCAAGCAGGGCCGAAUAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA-CCCUCGAGAGGUCACCGGGGUCCC--UGUCGGCCC----------- --..........(((((((...(((((.((.((((((.(((.....))))))))).))...........-.(((....)))......)))))..--..)))))))----------- ( -38.00, z-score = -2.02, R) >droSim1.chr3L 21981071 113 - 22553184 --CAACCAAGCAGGGCCGAACAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA-ACCUCGAGAGGUCACCGGGGUCCCGUCCUCUGUUCCCUGUCGGCCC --..........(((((((((((.....((.((((((.(((.....))))))))).))..)))).....-......(((((....(((....))).)))))........))))))) ( -43.60, z-score = -2.47, R) >droSec1.super_11 2614557 113 - 2888827 --CAACCAAGCAGGGCCGAACAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA-CCCUCGAGAGGUCACCGGGGUCCCGUCCUCUGUUCCCUGUCGGCCC --..........(((((((((((.....((.((((((.(((.....))))))))).))..)))).....-......(((((....(((....))).)))))........))))))) ( -43.60, z-score = -2.33, R) >droYak2.chr3L 23181874 108 - 24197627 --CAACCACCCAGGGCCGAACAGAUCCGACCUGAGCCAUUCCACCAGGAGGCUGAAGUGGCUGUAUUAA-CCCUCGAGAGGUCACC-GGGUCCCGUUCUCCGUUCCAUGUCG---- --..........((..((...(((...(((((((((((((.((((....)).)).))))))).......-.(((....)))....)-)))))....))).))..))......---- ( -29.70, z-score = 0.87, R) >droEre2.scaffold_4784 22345368 111 - 25762168 ACCAACCAACCAGGCCCGAACAGAUUCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUGGCUGUAUUAA-CCCUCGAGAGGUCACCGGGGUCCCGUUCCCUGUUCCCUGUUG---- ......((((.(((...((((((....((((((((((.(((.....))))))))..(.((.(......)-.)).)...)))))..(((....)))....)))))))))))))---- ( -37.10, z-score = -1.38, R) >droAna3.scaffold_13337 22435310 96 - 23293914 -----------CAAUUUGAUCCGAGCCGGCCUGAGCCGACAUAACGUGAGGUUAAAGUGGCUUUAUUAACCCCUUGAGAGGUCACGGGGGCCACAACCUCCUUUUCG--------- -----------..........((((.((((....)))).......(.((((((...(((((........((((.(((....))).))))))))))))))))..))))--------- ( -29.60, z-score = -1.12, R) >consensus __CAACCAAGCAGGGCCGAACAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA_CCCUCGAGAGGUCACCGGGGUCCCGUCCUCUGUUCCCUGUCG____ ............(((((..((((.....((.((((((.(((.....))))))))).))..)))).......(((....))).......)))))....................... (-20.19 = -20.78 + 0.59)
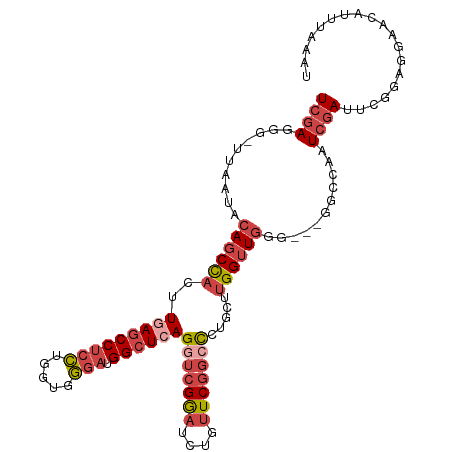
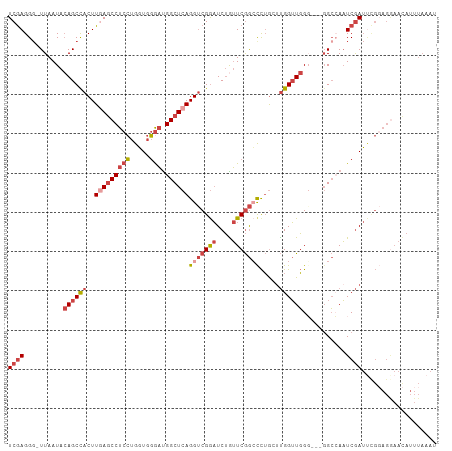
| Location | 22,694,571 – 22,694,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.42423 |
| G+C content | 0.51617 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -20.37 |
| Energy contribution | -22.07 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
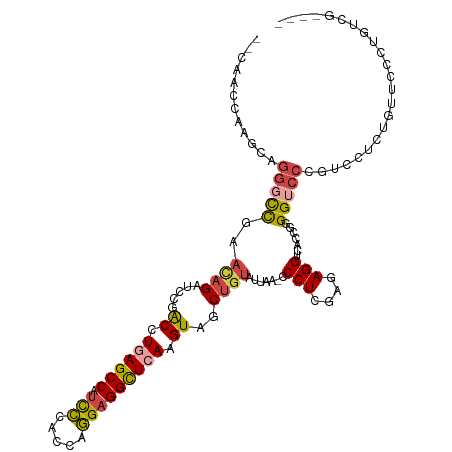
>dm3.chr3L 22694571 105 + 24543557 UCGAGGG-UUAAUACAGCUACUUGAGCCUCCUGGUGGGAUGGCUCAGGUCGGAUCUAUUCGGCCCUGCUUGGUUGGG---GGCCAAUCGAUUCGUAGGAAUAUUUAAAU ..(((.(-((..(((....((((((((((((.....))).))))))))).(((((.(((.((((((.(......)))---))))))).))))))))..))).))).... ( -38.80, z-score = -1.90, R) >droSim1.chr3L 21981110 105 + 22553184 UCGAGGU-UUAAUACAGCUACUUGAGCCUCCUGGUGGGAUGGCUCAGGUCGGAUCUGUUCGGCCCUGCUUGGUUGGG---GGCCAAUCGAUUCGGAGGAACAUUUAAAU .....((-((.........((((((((((((.....))).)))))))))((((((.(((.((((((.(......)))---))))))).))))))...))))........ ( -39.60, z-score = -1.84, R) >droSec1.super_11 2614596 105 + 2888827 UCGAGGG-UUAAUACAGCUACUUGAGCCUCCUGGUGGGAUGGCUCAGGUCGGAUCUGUUCGGCCCUGCUUGGUUGGG---GGCCAAUCGAUUCGGAGAAACAUUUAAAU (((((((-(..(.((((((((((((((((((.....))).))))))))).))..)))))..))))...((((((((.---..)))))))).)))).............. ( -38.30, z-score = -1.50, R) >droYak2.chr3L 23181908 103 + 24197627 UCGAGGG-UUAAUACAGCCACUUCAGCCUCCUGGUGGAAUGGCUCAGGUCGGAUCUGUUCGGCCCUGGGUGGUUGG-----GCGAAUCGAUUUGGAGGAACAUUUAAAU .....((-((.....)))).(((((((((((....)))..))))((((((((.((.((((((((......))))))-----)))).))))))))))))........... ( -33.30, z-score = -0.25, R) >droEre2.scaffold_4784 22345403 108 + 25762168 UCGAGGG-UUAAUACAGCCACUUGAGCCUCCUGGUGGGAUGGCUCAGGUCGAAUCUGUUCGGGCCUGGUUGGUUGGUUGUGGCGAAUCGAUAUGGAGGAACAUUUAAAU ((((.((-((.....)))).(((((((((((.....))).))))))))))))...(((((...((..(((((((.((....)).)))))))..))..)))))....... ( -37.90, z-score = -1.80, R) >droAna3.scaffold_13337 22435340 90 + 23293914 UCAAGGGGUUAAUAAAGCCACUUUAACCUCACGUUAUGUCGGCUCAGGCCGGCUCGGAUCAAAUUGAGUCGGGUCGGA--GGGCAUUCAAAC----------------- ....((((((((..........))))))))......((...((((...((((((((..((.....))..)))))))).--))))...))...----------------- ( -31.00, z-score = -3.27, R) >consensus UCGAGGG_UUAAUACAGCCACUUGAGCCUCCUGGUGGGAUGGCUCAGGUCGGAUCUGUUCGGCCCUGCUUGGUUGGG___GGCCAAUCGAUUCGGAGGAACAUUUAAAU ((((..........((((((..(((((((((.....))).))))))(((((((....))))))).....))))))...........))))................... (-20.37 = -22.07 + 1.70)

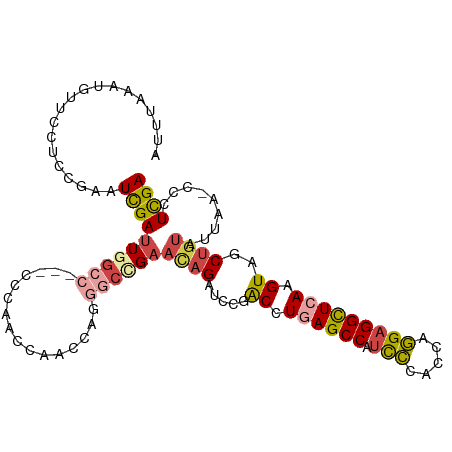
| Location | 22,694,571 – 22,694,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.42423 |
| G+C content | 0.51617 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.79 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22694571 105 - 24543557 AUUUAAAUAUUCCUACGAAUCGAUUGGCC---CCCAACCAAGCAGGGCCGAAUAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA-CCCUCGA .....(((((..(((((.(((..((((((---(...........)))))))...))).)....((((((.(((.....))))))))).))))..)))))..-....... ( -33.10, z-score = -3.77, R) >droSim1.chr3L 21981110 105 - 22553184 AUUUAAAUGUUCCUCCGAAUCGAUUGGCC---CCCAACCAAGCAGGGCCGAACAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA-ACCUCGA ...................((((((((((---(...........)))))))((((.....((.((((((.(((.....))))))))).))..)))).....-...)))) ( -32.10, z-score = -2.94, R) >droSec1.super_11 2614596 105 - 2888827 AUUUAAAUGUUUCUCCGAAUCGAUUGGCC---CCCAACCAAGCAGGGCCGAACAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA-CCCUCGA ...................((((((((((---(...........)))))))((((.....((.((((((.(((.....))))))))).))..)))).....-...)))) ( -32.10, z-score = -2.88, R) >droYak2.chr3L 23181908 103 - 24197627 AUUUAAAUGUUCCUCCAAAUCGAUUCGC-----CCAACCACCCAGGGCCGAACAGAUCCGACCUGAGCCAUUCCACCAGGAGGCUGAAGUGGCUGUAUUAA-CCCUCGA ...................((((.((((-----((.........))).)))((((..((.((.(.((((..(((....))))))).).)))))))).....-...)))) ( -23.50, z-score = -0.30, R) >droEre2.scaffold_4784 22345403 108 - 25762168 AUUUAAAUGUUCCUCCAUAUCGAUUCGCCACAACCAACCAACCAGGCCCGAACAGAUUCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUGGCUGUAUUAA-CCCUCGA ...................((((...(((((............(((..((((....)))).)))(((((.(((.....))))))))..))))).((....)-)..)))) ( -27.00, z-score = -2.52, R) >droAna3.scaffold_13337 22435340 90 - 23293914 -----------------GUUUGAAUGCCC--UCCGACCCGACUCAAUUUGAUCCGAGCCGGCCUGAGCCGACAUAACGUGAGGUUAAAGUGGCUUUAUUAACCCCUUGA -----------------...((((.((((--(..((((((..((.....))..)).(.((((....)))).).........))))..)).)))))))............ ( -17.90, z-score = -0.26, R) >consensus AUUUAAAUGUUCCUCCGAAUCGAUUGGCC___CCCAACCAACCAGGGCCGAACAGAUCCGACCUGAGCCAUCCCACCAGGAGGCUCAAGUAGCUGUAUUAA_CCCUCGA ...................((((((((((................))))))((((.....((.((((((.(((.....))))))))).))..)))).........)))) (-14.62 = -15.79 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:34 2011