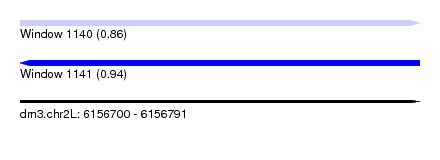
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,156,700 – 6,156,791 |
| Length | 91 |
| Max. P | 0.935313 |

| Location | 6,156,700 – 6,156,791 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 64.42 |
| Shannon entropy | 0.53753 |
| G+C content | 0.53384 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
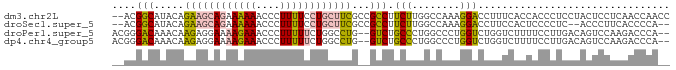
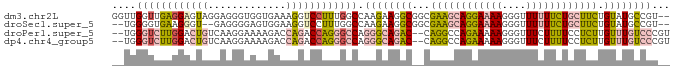
>dm3.chr2L 6156700 91 + 23011544 --ACGGCAUACAGAAGCAGAAAAAACCCUUUUCCUGCUUCGCCGCCUUCUUGGCCAAAGGACCUUUCACCACCCUCCUACUCCUCAACCAACC --..(((.....(((((((.((((....)))).))))))))))(((.....)))...((((.............))))............... ( -20.12, z-score = -3.40, R) >droSec1.super_5 4227670 87 + 5866729 --ACGGCAUACAGAAGCAGAAAAAACCCUUUUCCUGCUUCGCCGCCUUCUUGGCCAAAGGACCUUCCACUCCCCUC--ACCCUUCACCCCA-- --..(((.....(((((((.((((....)))).))))))))))(((.....)))....((.....)).........--.............-- ( -20.10, z-score = -3.09, R) >droPer1.super_5 5687 89 - 6813705 ACGGGACAAACAAGAGGAAAAGAAACCCUUUUUCUGGCCUG--GUCUGCCCUGGCCCUGGUCUGGUCUUUUCCUUGACAGUCCAAGACCCA-- ...((((......((((((((((...((....((.((((.(--(....))..))))..))...))))))))))))....))))........-- ( -29.20, z-score = -0.60, R) >dp4.chr4_group5 6350 89 - 2436548 ACGGGACAAACAAGAGGAAAAGAAACCCUUUUUCUGGCCUG--GUCUGCCCUGGCCCUGGUCUGGUCUUUUCCUUGACAGUCCAAGACCCA-- ...((((......((((((((((...((....((.((((.(--(....))..))))..))...))))))))))))....))))........-- ( -29.20, z-score = -0.60, R) >consensus __ACGACAAACAAAAGCAAAAAAAACCCUUUUCCUGCCCCG__GCCUGCCCGGCCAAAGGACCGGUCACUUCCCUCACACUCCAAAACCCA__ ....(((.....((((((((((((....))))))))))))...))).(((........)))................................ (-10.35 = -10.35 + -0.00)
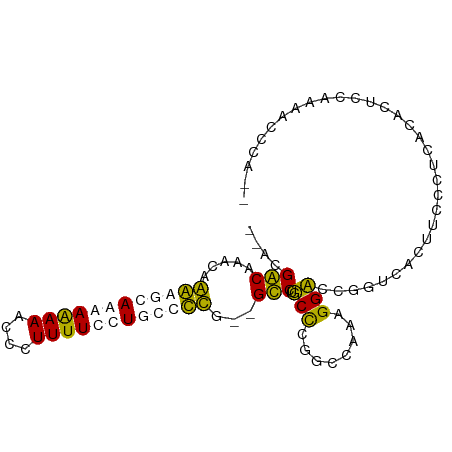
| Location | 6,156,700 – 6,156,791 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 64.42 |
| Shannon entropy | 0.53753 |
| G+C content | 0.53384 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -17.61 |
| Energy contribution | -19.05 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6156700 91 - 23011544 GGUUGGUUGAGGAGUAGGAGGGUGGUGAAAGGUCCUUUGGCCAAGAAGGCGGCGAAGCAGGAAAAGGGUUUUUUCUGCUUCUGUAUGCCGU-- ..(((((..((((.(...............).))).)..)))))...((((((((((((((((((....)))))))))))).)).))))..-- ( -30.66, z-score = -2.12, R) >droSec1.super_5 4227670 87 - 5866729 --UGGGGUGAAGGGU--GAGGGGAGUGGAAGGUCCUUUGGCCAAGAAGGCGGCGAAGCAGGAAAAGGGUUUUUUCUGCUUCUGUAUGCCGU-- --..........(((--.((((((.(....).)))))).))).....((((((((((((((((((....)))))))))))).)).))))..-- ( -32.90, z-score = -3.01, R) >droPer1.super_5 5687 89 + 6813705 --UGGGUCUUGGACUGUCAAGGAAAAGACCAGACCAGGGCCAGGGCAGAC--CAGGCCAGAAAAAGGGUUUCUUUUCCUCUUGUUUGUCCCGU --..((((((((.(((((........)).))).)))))))).((((((((--.(((..(((((.....)))))...)))...))))))))... ( -31.70, z-score = -0.56, R) >dp4.chr4_group5 6350 89 + 2436548 --UGGGUCUUGGACUGUCAAGGAAAAGACCAGACCAGGGCCAGGGCAGAC--CAGGCCAGAAAAAGGGUUUCUUUUCCUCUUGUUUGUCCCGU --..((((((((.(((((........)).))).)))))))).((((((((--.(((..(((((.....)))))...)))...))))))))... ( -31.70, z-score = -0.56, R) >consensus __UGGGUCGAGGACUGUCAAGGAAAAGAAAAGACCAGGGCCAAAGAAGAC__CAAACCAGAAAAAGGGUUUCUUCUCCUCCUGUAUGCCCC__ ....((((((((((((.............))))))))))))......((((..((((((((((((....))))))))))))....)))).... (-17.61 = -19.05 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:34 2011