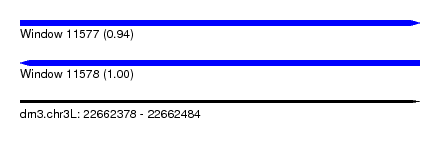
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,662,378 – 22,662,484 |
| Length | 106 |
| Max. P | 0.995555 |

| Location | 22,662,378 – 22,662,484 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.04 |
| Shannon entropy | 0.30491 |
| G+C content | 0.42230 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.65 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.937889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22662378 106 + 24543557 UGUGAUGUUAACGGGUG-UUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCAGA-UUGAAUUAAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAUAGACACGCCGAUC ...........((((((-((.....(((((((((((.((....(((((.....-............))))).....)).)))))))))))......))))).)))... ( -28.93, z-score = -2.45, R) >droSim1.chr3L 21947670 106 + 22553184 UGUGAUGUUAACGGGUG-UUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCAGA-UUGAAUUGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAGACACGCCGAUC ...........((((((-((.....(((((((((((.((....(((((.(((.-.....)))....))))).....)).)))))))))))......))))).)))... ( -30.20, z-score = -2.77, R) >droSec1.super_11 2578673 104 + 2888827 --UGAUGUUAACGGGUG-UUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCAGA-UUGAAUUGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAGACACGCCGAUC --.........((((((-((.....(((((((((((.((....(((((.(((.-.....)))....))))).....)).)))))))))))......))))).)))... ( -30.20, z-score = -3.13, R) >droYak2.chr3L 23128646 106 + 24197627 UUCGAUGUUAACGGGUG-UUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCAGA-UUGAGUUAAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAGACACGCCGAUC ...........((((((-((.....(((((((((((.((....(((((.....-............))))).....)).)))))))))))......))))).)))... ( -28.93, z-score = -2.32, R) >droEre2.scaffold_4784 22312270 106 + 25762168 UUUGAUGUAAACGGGUG-UUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCAGA-UUGAAUGGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAGCCACGCCGAUC ...........((((((-((.....(((((((((((.((....(((((.....-............))))).....)).))))))))))).....)).))).)))... ( -25.33, z-score = -1.11, R) >droAna3.scaffold_13337 22380397 106 + 23293914 AUCGGUGUUUACGGGUGUUUCUCGAAAGUAGAAUAGAAGAAUCUCACAGUGG--GUUAAAGGAACCUGUGAUAAGCCUGCUAUUCUACUUGCUAAAAACCCACCGGUC (((((((....((((.....)))).(((((((((((.((....((((((.(.--.(.....)..))))))).....)).)))))))))))..........))))))). ( -33.10, z-score = -2.75, R) >droWil1.scaffold_180698 4361932 108 - 11422946 UUUGUUGUUAUCGGGUGCUUUUCGAAAGUAGAAUAGAAGAAUCUCACACCGGUUGUAAAGGGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAGACACACCGUUA ...........((((((((((....(((((((((((.((....(((((...(((.(....).))).))))).....)).)))))))))))...)))).))).)))... ( -29.50, z-score = -1.87, R) >droGri2.scaffold_15110 15103984 106 + 24565398 UGUGACGCUAGCUGGUGUUUGUCGAAAGUAGAAUAGAAGAAUCUCACACCAUG-UCGAAUGUUACAUGUGAUAAGCCUGCUAUUCUACUUGCCA-AUCCACACCGAUC ............((((((..((.(.(((((((((((.((....(((((...((-(........)))))))).....)).)))))))))))..).-))..))))))... ( -25.10, z-score = -1.07, R) >droMoj3.scaffold_6680 2679265 103 - 24764193 ---GACGCUGACUGGUGCUUGUCGAAAGUAGAAUAGAAGAAUCUCACACUCGG-UCAAAUGGUACAUGUGAUAAGCCUGCUAUUCUACUUGCCA-AUCCACACCGACU ---.........(((((........(((((((((((.((....(((((.(((.-.....)))....))))).....)).)))))))))))....-.....)))))... ( -22.53, z-score = -0.23, R) >droVir3.scaffold_13049 8143642 103 - 25233164 ---GACGCUAGCUGGUGUUUGUCGAAAGUAGAAUAGAAGAAUCUCACACUAGG-UUGAAUUGUACAUGUGAUAAGCCUGCUAUUCUACUUGCCA-AUCCACACCGAUC ---.........((((((..((.(.(((((((((((.((....(((((....(-(........)).))))).....)).)))))))))))..).-))..))))))... ( -25.10, z-score = -1.16, R) >droPer1.super_96 69850 103 - 202244 ----AUGUCAACGGGUGUUUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCGGA-UUGAAUGGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAUACACACCGAUC ----.......(((((((.......(((((((((((.((....(((((..(..-..........).))))).....)).))))))))))).......)))).)))... ( -27.74, z-score = -2.06, R) >dp4.chrXR_group8 344175 103 + 9212921 ----AUGUCAACGGGUGUUUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCGGA-UUGAAUGGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAUACACACCGAUC ----.......(((((((.......(((((((((((.((....(((((..(..-..........).))))).....)).))))))))))).......)))).)))... ( -27.74, z-score = -2.06, R) >consensus U_UGAUGUUAACGGGUG_UUGUCGAAAGUAGAAUAGAAGAAUCUCACAGCAGA_UUGAAUGGAACAUGUGAUAAGCCUGCUAUUCUACUUGCCAAAGACACACCGAUC .............((((........(((((((((((.((....(((((..................))))).....)).)))))))))))..........)))).... (-21.89 = -21.65 + -0.24)

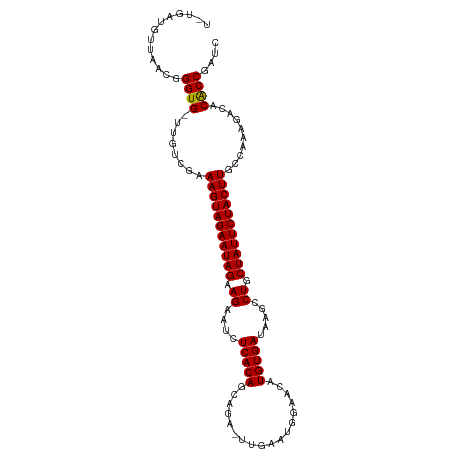
| Location | 22,662,378 – 22,662,484 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Shannon entropy | 0.30491 |
| G+C content | 0.42230 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22662378 106 - 24543557 GAUCGGCGUGUCUAUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUUAAUUCAA-UCUGCUGUGAGAUUCUUCUAUUCUACUUUCGACAA-CACCCGUUAACAUCACA ...(((.((((...(((.(((((((((((.(((....(((((.((.........-...)))))))...))).))))))))))))))...)-))))))........... ( -29.00, z-score = -3.10, R) >droSim1.chr3L 21947670 106 - 22553184 GAUCGGCGUGUCUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCAAUUCAA-UCUGCUGUGAGAUUCUUCUAUUCUACUUUCGACAA-CACCCGUUAACAUCACA ...(((.((((..((((.(((((((((((.(((....(((((.((.........-...)))))))...))).)))))))))))))))..)-))))))........... ( -31.70, z-score = -4.03, R) >droSec1.super_11 2578673 104 - 2888827 GAUCGGCGUGUCUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCAAUUCAA-UCUGCUGUGAGAUUCUUCUAUUCUACUUUCGACAA-CACCCGUUAACAUCA-- ...(((.((((..((((.(((((((((((.(((....(((((.((.........-...)))))))...))).)))))))))))))))..)-)))))).........-- ( -31.70, z-score = -4.10, R) >droYak2.chr3L 23128646 106 - 24197627 GAUCGGCGUGUCUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUUAACUCAA-UCUGCUGUGAGAUUCUUCUAUUCUACUUUCGACAA-CACCCGUUAACAUCGAA ...(((.((((..((((.(((((((((((.(((....(((((.((.........-...)))))))...))).)))))))))))))))..)-))))))........... ( -31.70, z-score = -3.66, R) >droEre2.scaffold_4784 22312270 106 - 25762168 GAUCGGCGUGGCUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCCAUUCAA-UCUGCUGUGAGAUUCUUCUAUUCUACUUUCGACAA-CACCCGUUUACAUCAAA ...(((.(((...((((.(((((((((((.(((....(((((.((.........-...)))))))...))).)))))))))))))))...-))))))........... ( -29.90, z-score = -3.04, R) >droAna3.scaffold_13337 22380397 106 - 23293914 GACCGGUGGGUUUUUAGCAAGUAGAAUAGCAGGCUUAUCACAGGUUCCUUUAAC--CCACUGUGAGAUUCUUCUAUUCUACUUUCGAGAAACACCCGUAAACACCGAU ...(((((.((((((.(.(((((((((((.(((....((((((...........--...))))))...))).))))))))))).).)))))).........))))).. ( -34.14, z-score = -4.05, R) >droWil1.scaffold_180698 4361932 108 + 11422946 UAACGGUGUGUCUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCCCUUUACAACCGGUGUGAGAUUCUUCUAUUCUACUUUCGAAAAGCACCCGAUAACAACAAA ...(((.(((..(((.(.(((((((((((.(((....(((((((((........)))..))))))...))).))))))))))).).)))..))))))........... ( -34.40, z-score = -4.40, R) >droGri2.scaffold_15110 15103984 106 - 24565398 GAUCGGUGUGGAU-UGGCAAGUAGAAUAGCAGGCUUAUCACAUGUAACAUUCGA-CAUGGUGUGAGAUUCUUCUAUUCUACUUUCGACAAACACCAGCUAGCGUCACA ....(((((...(-(((.(((((((((((.((.(((((..(((((........)-))))..)))))...)).)))))))))))))))...)))))............. ( -34.50, z-score = -3.24, R) >droMoj3.scaffold_6680 2679265 103 + 24764193 AGUCGGUGUGGAU-UGGCAAGUAGAAUAGCAGGCUUAUCACAUGUACCAUUUGA-CCGAGUGUGAGAUUCUUCUAUUCUACUUUCGACAAGCACCAGUCAGCGUC--- ....(((((...(-(((.(((((((((((.(((....((((((...........-....))))))...))).)))))))))))))))...)))))..........--- ( -31.46, z-score = -2.03, R) >droVir3.scaffold_13049 8143642 103 + 25233164 GAUCGGUGUGGAU-UGGCAAGUAGAAUAGCAGGCUUAUCACAUGUACAAUUCAA-CCUAGUGUGAGAUUCUUCUAUUCUACUUUCGACAAACACCAGCUAGCGUC--- ....(((((...(-(((.(((((((((((.(((....((((((...........-....))))))...))).)))))))))))))))...)))))..........--- ( -31.16, z-score = -2.85, R) >droPer1.super_96 69850 103 + 202244 GAUCGGUGUGUAUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCCAUUCAA-UCCGCUGUGAGAUUCUUCUAUUCUACUUUCGACAAACACCCGUUGACAU---- ...(((.((((..((((.(((((((((((.(((....(((((.((.........-...)))))))...))).)))))))))))))))...))))))).......---- ( -29.30, z-score = -2.88, R) >dp4.chrXR_group8 344175 103 - 9212921 GAUCGGUGUGUAUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCCAUUCAA-UCCGCUGUGAGAUUCUUCUAUUCUACUUUCGACAAACACCCGUUGACAU---- ...(((.((((..((((.(((((((((((.(((....(((((.((.........-...)))))))...))).)))))))))))))))...))))))).......---- ( -29.30, z-score = -2.88, R) >consensus GAUCGGUGUGUCUUUGGCAAGUAGAAUAGCAGGCUUAUCACAUGUUCCAUUCAA_UCUGCUGUGAGAUUCUUCUAUUCUACUUUCGACAA_CACCCGUUAACAUCA_A .......(((....(.(.(((((((((((.(((....(((((..................)))))...))).))))))))))).).)....))).............. (-19.91 = -20.00 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:27 2011