
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,646,239 – 22,646,336 |
| Length | 97 |
| Max. P | 0.597412 |

| Location | 22,646,239 – 22,646,336 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Shannon entropy | 0.35093 |
| G+C content | 0.50272 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22646239 97 + 24543557 ------------CGAAAGCACUGGGAGGUGCGGGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUCAUGG-- ------------...........((((((((.(((((..(((....)))...))))).....((((...((((((((....))))))))....))))))))))))....-- ( -28.10, z-score = -0.98, R) >droSim1.chr3L 21930431 97 + 22553184 ------------CGAGAGCAGUGGGAGGAGCGGGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUCAUGG-- ------------...(((.(((((.((.(((..(.(((......))).)..))).)).)..(((((...((((((((....))))))))....))))))))))))....-- ( -28.60, z-score = -0.86, R) >droSec1.super_11 2562622 97 + 2888827 ------------CGAGAGCAGUGGGAGGAGCGGGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUCAUGG-- ------------...(((.(((((.((.(((..(.(((......))).)..))).)).)..(((((...((((((((....))))))))....))))))))))))....-- ( -28.60, z-score = -0.86, R) >droYak2.chr3L 23102060 97 + 24197627 ------------CGAGAGCAGUGGGAGGAGCGGGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUCAUGG-- ------------...(((.(((((.((.(((..(.(((......))).)..))).)).)..(((((...((((((((....))))))))....))))))))))))....-- ( -28.60, z-score = -0.86, R) >droEre2.scaffold_4784 22296401 97 + 25762168 ------------CGAGAGCAGUGGGAGGAGCGGGCGGAAAUGACUCCAUAAGCUGCUGCUACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUCAUGG-- ------------.(((((((((..(.((((((........)).)))).)..)))))((((((.((....((((((((....))))))))...)))))))).))))....-- ( -30.50, z-score = -1.39, R) >droAna3.scaffold_13337 22360801 97 + 23293914 ------------AAGGAGCAGUGGGAGGGAU-GGCGGAAAUGACUUCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUUCAUGG- ------------..(.((((((.((((..((-.......))..))))....)))))).)..(((((...((((((((....))))))))....)))))............- ( -27.20, z-score = -1.06, R) >droWil1.scaffold_180698 7091486 111 + 11422946 UGGAACCUGAGGCAGCCGAAGAGCGGGACGUAAACGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUUUCAUGG .....(((((((((((....(((((...((....))....)).))).....))))))....(((((...((((((((....))))))))....))))).......))).)) ( -28.10, z-score = -0.30, R) >droVir3.scaffold_13049 17108705 84 + 25233164 -----------------------AGACGAGCCAGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUUGCUGAUGUGGCAGUUUUCG---- -----------------------...(((......(((......)))..(((((((..((..((.....((((((((....)))))))).)).))..))))))))))---- ( -20.90, z-score = -0.79, R) >droMoj3.scaffold_6680 22109605 84 + 24764193 -----------------------AGACGAGCCAGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUUGCUGAUGUGGCAGUUUUCG---- -----------------------...(((......(((......)))..(((((((..((..((.....((((((((....)))))))).)).))..))))))))))---- ( -20.90, z-score = -0.79, R) >droGri2.scaffold_15110 15083549 84 + 24565398 -----------------------AGACGAGCCAGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUUGCUGAUGUGGCACUUUUCG---- -----------------------.((.(((.((((((..(((....)))...))))))...(((((...((((((((....))))))))....)))))..))).)).---- ( -21.20, z-score = -1.29, R) >consensus ____________CGAGAGCAGUGGGAGGAGCGGGCGGAAAUGACUCCAUAAGCUGCUGCAACCAUAAACCGGAAGUUAACAGACUUUCGCUGAUGUGGCACUUUCAUGG__ ......................(((((..((.(((((..(((....)))...)))))))..(((((...((((((((....))))))))....)))))..)))))...... (-21.79 = -21.23 + -0.56)

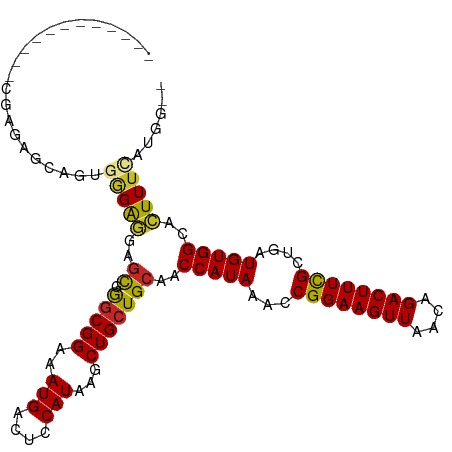
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:23 2011