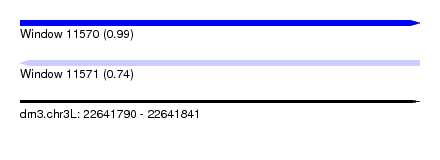
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,641,790 – 22,641,841 |
| Length | 51 |
| Max. P | 0.991713 |

| Location | 22,641,790 – 22,641,841 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 65.61 |
| Shannon entropy | 0.62191 |
| G+C content | 0.49566 |
| Mean single sequence MFE | -14.57 |
| Consensus MFE | -10.43 |
| Energy contribution | -9.88 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
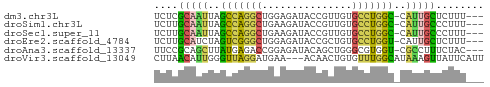
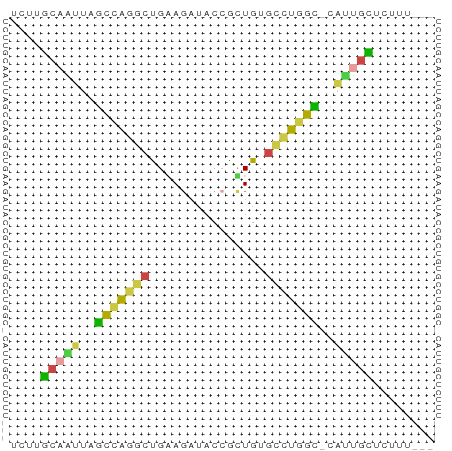
>dm3.chr3L 22641790 51 + 24543557 UCUCGCAAUUAGCCAGGCUGGAGAUACCGUUGUGCCUGGC-CAUUGCUCUUU--- ....(((((..((((((((((.....)))....)))))))-.))))).....--- ( -19.40, z-score = -2.55, R) >droSim1.chr3L 21925484 51 + 22553184 UCUUGCAAUUAGCCAGGCUGAAGAUACCGUUGUGCCUGGC-CAUUGCCCUUU--- ....(((((..(((((((...............)))))))-.))))).....--- ( -17.66, z-score = -2.73, R) >droSec1.super_11 2558211 51 + 2888827 UCUUGCAAUUAGCCAGGCUGAAGAUACCGUUGUGCCUGGC-CAUUGCCCUUU--- ....(((((..(((((((...............)))))))-.))))).....--- ( -17.66, z-score = -2.73, R) >droEre2.scaffold_4784 22292033 51 + 25762168 UCUUGCAUCUAGUCGGGCUGGAGAUACCGCUGUGCCUGGU-CAUUGCUCUUU--- ....(((..(..(((((((((.(....).))).)))))).-.).))).....--- ( -11.00, z-score = 0.73, R) >droAna3.scaffold_13337 22355865 51 + 23293914 UUCCGCAGCUUAUGAGACCGGAGAUACAGCUGGGCGUGGU-CGCCUUUCUAC--- (((((...((....))..)))))....((..(((((....-)))))..))..--- ( -13.40, z-score = -0.05, R) >droVir3.scaffold_13049 8118755 52 - 25233164 CUUAACAUUGGGUUAGGAUGAA---ACAACUGUGUUUGGCAUAAAGUUAUUCAUU ((((((.....))))))(((((---..(((((((.....)))..)))).))))). ( -8.30, z-score = -0.66, R) >consensus UCUUGCAAUUAGCCAGGCUGAAGAUACCGCUGUGCCUGGC_CAUUGCUCUUU___ ....(((((..(((((((...............)))))))..)))))........ (-10.43 = -9.88 + -0.55)
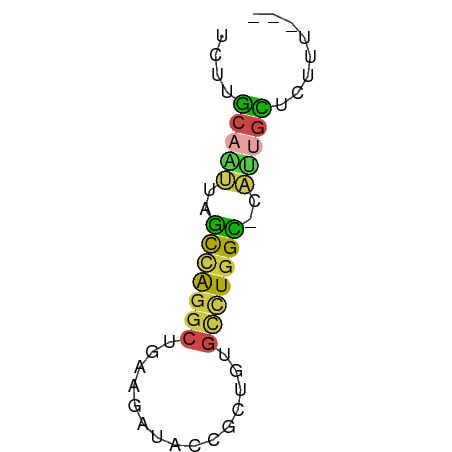
| Location | 22,641,790 – 22,641,841 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 65.61 |
| Shannon entropy | 0.62191 |
| G+C content | 0.49566 |
| Mean single sequence MFE | -14.00 |
| Consensus MFE | -5.82 |
| Energy contribution | -6.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 22641790 51 - 24543557 ---AAAGAGCAAUG-GCCAGGCACAACGGUAUCUCCAGCCUGGCUAAUUGCGAGA ---.....((((((-(((((((.....((.....)).)))))))).))))).... ( -21.50, z-score = -3.37, R) >droSim1.chr3L 21925484 51 - 22553184 ---AAAGGGCAAUG-GCCAGGCACAACGGUAUCUUCAGCCUGGCUAAUUGCAAGA ---.....((((((-(((((((...............)))))))).))))).... ( -20.66, z-score = -3.11, R) >droSec1.super_11 2558211 51 - 2888827 ---AAAGGGCAAUG-GCCAGGCACAACGGUAUCUUCAGCCUGGCUAAUUGCAAGA ---.....((((((-(((((((...............)))))))).))))).... ( -20.66, z-score = -3.11, R) >droEre2.scaffold_4784 22292033 51 - 25762168 ---AAAGAGCAAUG-ACCAGGCACAGCGGUAUCUCCAGCCCGACUAGAUGCAAGA ---.....((..((-......))..)).((((((..((.....)))))))).... ( -6.10, z-score = 1.37, R) >droAna3.scaffold_13337 22355865 51 - 23293914 ---GUAGAAAGGCG-ACCACGCCCAGCUGUAUCUCCGGUCUCAUAAGCUGCGGAA ---.(((...((((-....))))...)))..(((.((((.......)))).))). ( -11.40, z-score = 0.36, R) >droVir3.scaffold_13049 8118755 52 + 25233164 AAUGAAUAACUUUAUGCCAAACACAGUUGU---UUCAUCCUAACCCAAUGUUAAG .((((((((((...((.....)).))))).---)))))................. ( -3.70, z-score = -0.13, R) >consensus ___AAAGAGCAAUG_GCCAGGCACAACGGUAUCUCCAGCCUGACUAAUUGCAAGA ........(((((....(((((...............)))))....))))).... ( -5.82 = -6.46 + 0.64)

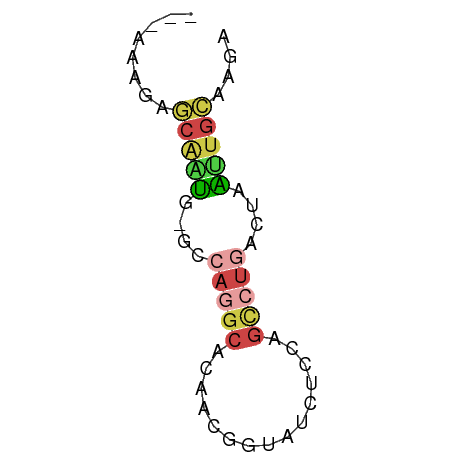
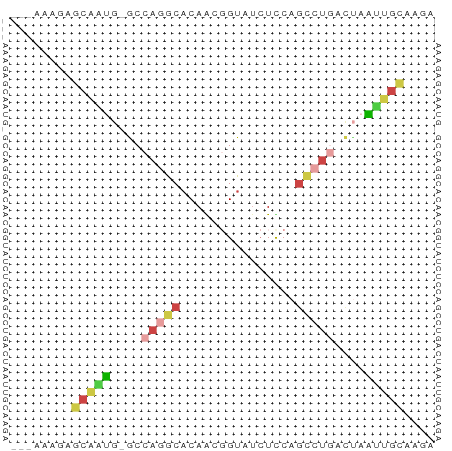
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:22 2011