
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 22,635,547 – 22,635,638 |
| Length | 91 |
| Max. P | 0.634377 |

| Location | 22,635,547 – 22,635,638 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 58.10 |
| Shannon entropy | 0.63049 |
| G+C content | 0.44429 |
| Mean single sequence MFE | -15.05 |
| Consensus MFE | -3.59 |
| Energy contribution | -4.35 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
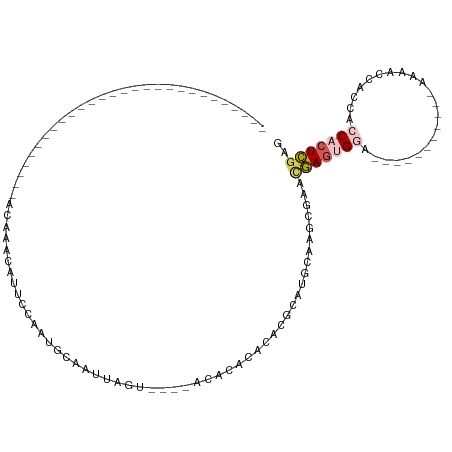
>dm3.chr3L 22635547 91 + 24543557 ------------GAUAUAAUAACAA--CAAAAACAUUCCAAUGCAAUUAGUUCACACGCACACACGCGUGCAAGUGAACGAGUGUA-----------AAAACUACCACCACUCGAG ------------.............--......................((((((..((((......))))..))))))(((((..-----------...........)))))... ( -18.92, z-score = -3.10, R) >droYak2.chr3L 23090613 69 + 24197627 --------------------------------GCAUUCCCAAGCAAUUAGU----UCACACACACAUAUGCAAGUGAACGAGUGGA-----------AAAACCACCACCACUCGAG --------------------------------((........)).....((----((((..............))))))((((((.-----------..........))))))... ( -15.94, z-score = -3.66, R) >droEre2.scaffold_4784 22286015 72 + 25762168 -----------------------------CAUACAUUCCCAUACAAUUAGU----UCACACACACACGUGCAAGCGAACGAGUGGA-----------AAAACCACCACCACUCGAG -----------------------------....................((----(..(((......)))..)))...(((((((.-----------..........))))))).. ( -14.60, z-score = -3.60, R) >droAna3.scaffold_13337 22350026 95 + 23293914 GUAACAAUAAUAACCACAGCGGCGAGACAGAAACAUUCCAAUGCAAUUAGU----GCGCACAUAAGCCUG------AAUGAGCGGA-----------AAAACCACCACCACUCGAG .............((.....))((((.......(((((....(((.....)----))((......))..)------))))...((.-----------....)).......)))).. ( -12.90, z-score = 0.14, R) >droVir3.scaffold_13049 8112239 89 - 25233164 -------------------------GAUACCAGCAUUCCAAUGCAGUUAGU--GCACACAGGCAUGCAUGCACACACACAAACGUAUUUACAUAAAUAAAACCACCACCACUUAAG -------------------------(((((..((((....)))).....((--(((..((....))..)))))..........)))))............................ ( -12.90, z-score = -1.08, R) >consensus ____________________________ACAAACAUUCCAAUGCAAUUAGU____ACACACACACGCAUGCAAGCGAACGAGUGGA___________AAAACCACCACCACUCGAG ..............................................................................(((((((......................))))))).. ( -3.59 = -4.35 + 0.76)

| Location | 22,635,547 – 22,635,638 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 58.10 |
| Shannon entropy | 0.63049 |
| G+C content | 0.44429 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -7.22 |
| Energy contribution | -6.46 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
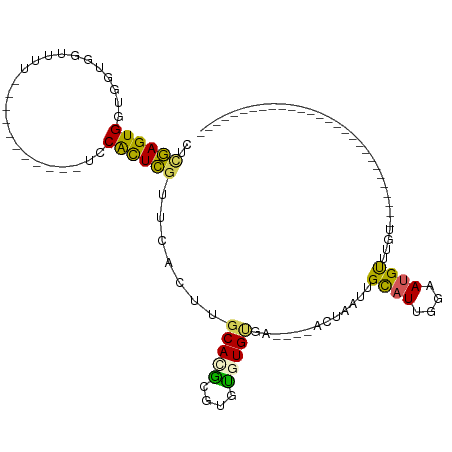
>dm3.chr3L 22635547 91 - 24543557 CUCGAGUGGUGGUAGUUUU-----------UACACUCGUUCACUUGCACGCGUGUGUGCGUGUGAACUAAUUGCAUUGGAAUGUUUUUG--UUGUUAUUAUAUC------------ ...(((((...........-----------..)))))((((((.((((((....)))))).))))))(((..((((....))))..)))--.............------------ ( -24.72, z-score = -1.76, R) >droYak2.chr3L 23090613 69 - 24197627 CUCGAGUGGUGGUGGUUUU-----------UCCACUCGUUCACUUGCAUAUGUGUGUGUGA----ACUAAUUGCUUGGGAAUGC-------------------------------- ((((((..(((((((....-----------.))))).((((((.((((....)))).))))----))..))..)))))).....-------------------------------- ( -25.60, z-score = -4.28, R) >droEre2.scaffold_4784 22286015 72 - 25762168 CUCGAGUGGUGGUGGUUUU-----------UCCACUCGUUCGCUUGCACGUGUGUGUGUGA----ACUAAUUGUAUGGGAAUGUAUG----------------------------- (((((((((..(.....).-----------.))))))((((((.((((....)))).))))----)).........)))........----------------------------- ( -21.30, z-score = -2.33, R) >droAna3.scaffold_13337 22350026 95 - 23293914 CUCGAGUGGUGGUGGUUUU-----------UCCGCUCAUU------CAGGCUUAUGUGCGC----ACUAAUUGCAUUGGAAUGUUUCUGUCUCGCCGCUGUGGUUAUUAUUGUUAC ....(((((((((((....-----------.)))).((((------(..((......))((----(.....)))....))))).........)))))))................. ( -23.00, z-score = -0.36, R) >droVir3.scaffold_13049 8112239 89 + 25233164 CUUAAGUGGUGGUGGUUUUAUUUAUGUAAAUACGUUUGUGUGUGUGCAUGCAUGCCUGUGUGC--ACUAACUGCAUUGGAAUGCUGGUAUC------------------------- .......((((.((((.((....(((((.(((((....)))))(((((((((....)))))))--))....)))))...)).)))).))))------------------------- ( -20.70, z-score = 0.34, R) >consensus CUCGAGUGGUGGUGGUUUU___________UCCACUCGUUCACUUGCACGCGUGUGUGUGA____ACUAAUUGCAUUGGAAUGUUUGU____________________________ ..((((((........................)))))).......(((((....))))).............((((....))))................................ ( -7.22 = -6.46 + -0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:45:20 2011